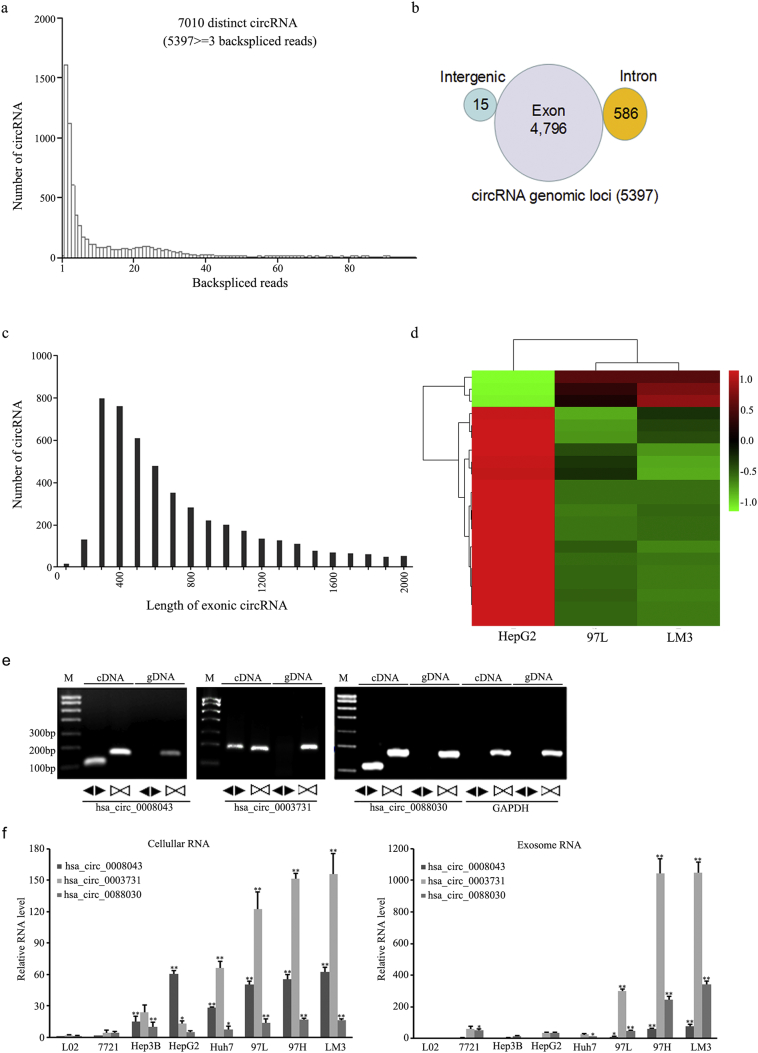
Fig. 4.
Identification of circRNAs in RNA sequencing data and the determination of differentially expressed circRNAs in HepG2, 97 L, and LM3 cell-derived exosomes. (a) The length distributions of backspliced circRNAs in HepG2, 97 L, and LM3 cells. (b) The genomic origins of HepG2, 97 L, and LM3 cell-derived circRNAs. (c) The length distributions of exonic circRNAs in HepG2, 97 L, and LM3 cells. (d) Heat map showing the expression profiles of exosomal circRNA in HepG2, 97 L, and LM3 cells. (e) The identification of three isoforms of circPTGR1 by RT-PCR, using divergent primers. (f) Fold enrichment of the three isoforms of circPTGR1 detected by qRT-PCR compared to the linear PTGR1 mRNAs in exosomes and their parent cells in eight hepatocellular carcinoma cell lines. Error bars indicate standard deviations. * P < .05 vs. L-O2; ** P < .01 vs. L-O2. (For cellular RNA, Hsa_circ_0008043: L-O2 vs. Hep3B, P = .0004; L-O2 vs. HepG2, huh7, 97 L, 97H, and LM3, P < .0001; Hsa_circ_0003731: L-O2 vs. huh, 97 L, 97H, hand LM3, P < .0001; Hsa_circ_0088030, L-O2 vs. Hep3B, P = .0038; L-O2 vs. huh7, P = .0397; L-O2 vs. 97 L, 97H, and LM3, P < .0001. For exosomal RNA, Hsa_circ_0008043: L-O2 vs. 97 L, P = .0435; L-O2 vs. 97H and LM3, P < .0001; Hsa_circ_0003731: L-O2 vs. huh, 97 L, 97H, and LM3, P < .0001; Hsa_circ_0088030, L-O2 vs. Hep3B, P = .0169; L-O2 vs. huh7, P = .0003; L-O2 vs. 97 L, P = .0007; L-O2 vs. 97H, LM3, P < .0001).