Figure 3.
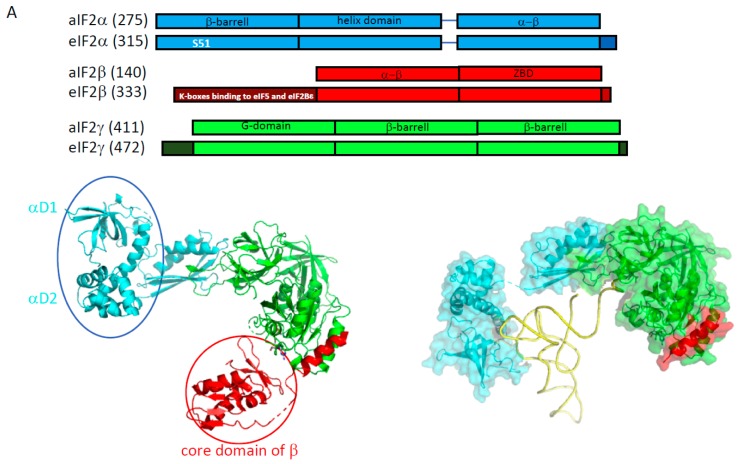
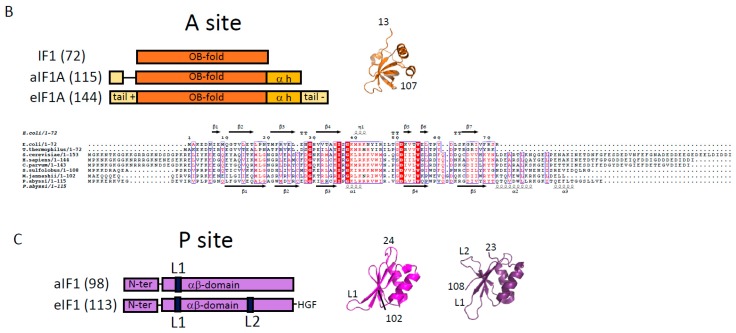
The structure of the initiation factors belonging to the core complex. (A) The structural organization of the three subunits of e/aIF2 is shown. Overall, aIF2α, β and γ from P. abyssi have about 28%, 40%, and 48% sequence identity with the eukaryotic eIF2α, β and γ subunits, respectively. The eukaryotic specificities of each factor are schematized by dark boxes or indicated within the block with white letters. The eukaryotic eIF2α have an acidic C-terminal extension. The eukaryotic eIF2β have a supplementary N-terminal domain necessary for the binding to eIF5 and eIF2Bε. The eukaryotic eIF2γ have N- and C-terminal extensions compared to the archaeal versions. Note that some eukaryotic and/or archaeal specificities are also found at some positions in the sequences (see text and [66,67,68] for sequence alignments). The structure of the full heterotrimeric aIF2 is shown in a cartoon representation in its unbound form (left, PDB ID 2AHO and 2QMU) and bound to Met-tRNAiMet (right, PDB ID 3V11). The α subunit is colored in cyan, the β subunit is colored in red and the γ subunit is colored in green. The two mobile wings of aIF2, the αD12 domains and the core domain of β, are circled. (B) The structures of e/aIF1A and of its bacterial homologue IF1. The structure of aIF1A from P. abyssi (PDB ID 4MNO) is drawn in orange cartoons. Overall, eIF1A from P. abyssi has about 40% identity with the eukaryotic eIF1A. The basic and acidic eukaryotic extensions necessary for long-range scanning are shown with light yellow boxes. The archaeal version of the factor contains a helix domain at the C-terminal extremity but no acidic extension. However, a short N-terminal tail is present in aIF1A. The organization of the bacterial IF1 protein is also shown. As described in the text, because of their structural resemblance, the IF1 and e/aIF1A proteins are considered as universal initiation factors. The secondary structures are from E. coli IF1 (PDB code 1AH9) and from P. abyssi aIF1A (PDB code 4MNO). (C) Structure of e/aIF1. The structure of aIF1 from Methanocaldococcus jannaschii (PDB ID 4MO0) is drawn in magenta cartoons, and that of eIF1 from S. cerevisiae (PDB ID 2OGH) is in purple. Overall, eIF1 from P. abyssi has about 30% sequence identity with the eukaryotic eIF1. The L1 loop, also called a basic loop, is located in front of the anticodon loop of the tRNA in the PIN states of eukaryotes and archaea. The loop is mobile outside of the ribosome (Figure 4). The L2 loop is only present in eukaryotes [72]. It contacts the tRNA D-loop in the PIN state [73]. In all the panels, the numbers of residues indicated in parentheses refer to P. abyssi proteins for the archaea, human proteins for eukaryotes, and E. coli for IF1.