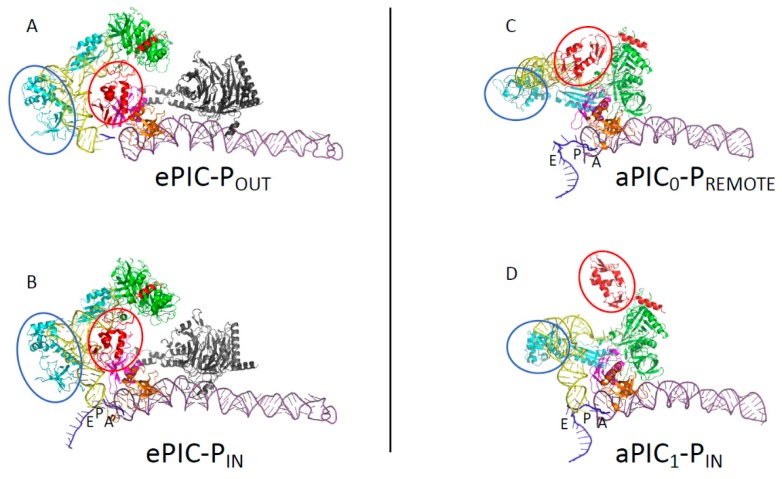
Figure 4.
A comparison of archaeal and eukaryotic initiation complexes. The figure shows partial views (P site region) of eukaryotic and archaeal initiation complexes. (A) The eukaryotic 48S-open POUT complex. (B) The eukaryotic 48S-closed PIN complex. (A) and (B) are from [134] (PDB ID 3JAQ, 3JAP). (C) The archaeal PIC0-PREMOTE conformation. (D) The archaeal PIC1-PIN conformation. The color code for the initiation factors is the same as in Figure 3. The small ribosomal helix h44 is in dark purple and the mRNA is in dark blue. The eIF3 subunits in views (A) and (B) are in black. The two mobile wings of e/aIF2, as defined in the legend of Figure 3, are encircled. Archaeal aIF1 and aIF1A have positions similar to those of their eukaryotic orthologues eIF1 and eIF1A. In eukaryotes, the initiator tRNA is bound to the γ subunit of eIF2 and to the domain 3 of eIF2α, as observed in the archaeal TC [69]. However, eIF2αD12 has moved and is found in the E site, while the core domain of β is close to the tRNA. No interaction is observed between the γ subunit and h44. In archaea, the structure of the TC bound to the ribosome corresponds to that observed outside the ribosome for the IC0-PREMOTE complex. The γ subunit contacts h44 and aIF1. In the IC1-PIN conformation, the structure of the TC is constrained, but the energetic cost would be compensated by the codon:anticodon base pairing. The movement of the two mobile wings of aIF2 may help the start codon selection.