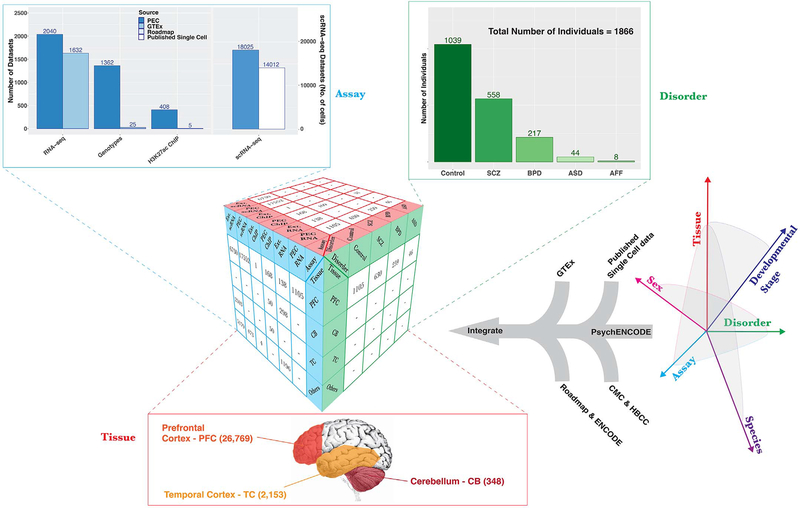
Fig. 1. Comprehensive data resource for functional genomics of the human brain.
The functional genomics data generated by the PsychENCODE Consortium (PEC) constitute a multidimensional exploration across tissue, developmental stage, disorder, species, assay, and sex. The central data cube represents the results of our data integration for the three dimensions of disorder, assay, and tissue, where the numbers of datasets in the analysis are depicted. Projections of the data onto each of these three parameters are shown as graphs for assay and disorder and as a schematic for the primary brain regions of interest. Assay: Dataset numbers for a subset of assays are shown, including RNA-seq (2040 PsychENCODE samples and 1632 GTEx samples, used in multiple downstream analyses), genotypes (1362 PsychENCODE and 25 GTEx individuals for a total of 1387 individuals matched to RNA-seq samples for QTL analysis after quality control filtering), and H3K27ac ChIP-seq (408 PsychENCODE and 5 Roadmap samples). The number of cells assayed by small conditional RNA sequencing (scRNA-seq) (right-hand y axis) is 18,025 for PsychENCODE and 14,012 for external (ext.) datasets. Disorder: Across all assays, there are 113 GTEx and 926 PsychENCODE control individuals and 558 SCZ, 217 BPD, 44 ASD, and 8 affective disorder (AFF) individuals from PsychENCODE, resulting in 1866 individuals. Tissue: Three brain regions are considered—the PFC (n = 26,769 samples), TC (n = 2153 samples), and CB (n = 348 samples). See table S11 and (19) for more details. HBCC, Human Brain Collection Core.