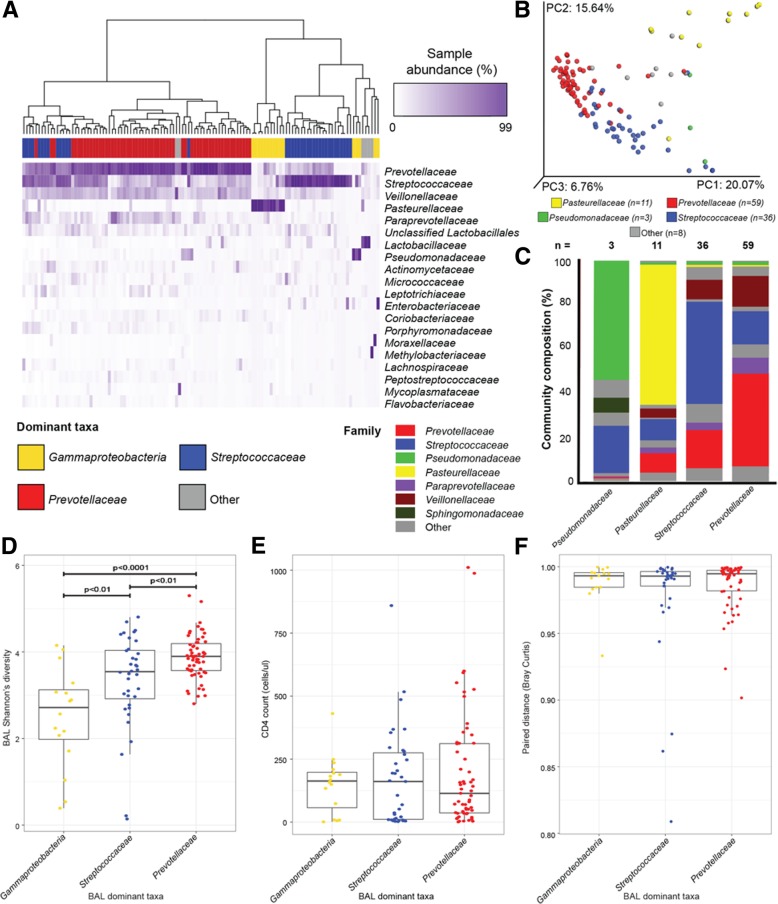
Fig. 2.
Three distinct airway microbiota exist in HIV-infected pneumonia patients and relate to microbiological factors, but not to peripheral CD4 count or gut microbiota composition. a Unsupervised hierarchical clustering (using Bray–Curtis dissimilarity (BC) and Ward 2 clustering) of BAL microbiota and abundance heat map of the top 20 bacterial families present in at least one sample at ≥ 3% relative abundance (ordered from highest to lowest abundance) indicate distinct but repeating patterns of microbial co-colonization in the lower airways of HIV-infected pneumonia patients. b Principal coordinates analysis (PCoA) of weighted UniFrac distance for n = 117 BAL samples confirms that significantly distinct airway microbiota co-associate with dominant bacterial family and explain a large proportion of taxonomic variation in lower airway bacterial communities (PERMANOVA, R2 = 0.658, p < 0.001). c Mean community composition at the family level for each of the distinct microbiota community states. Bacterial community states differ in d Shannon’s diversity (KW, p < 0.0001; Mann–Whitney U test p values plotted), but do not relate to e peripheral CD4 count (cells μl−1; KW p = 0.85) or to f similarity to paired stool sample (paired distance measured using Bray–Curtis; KW, p = 0.62). PC, principal coordinate; PERMANOVA, permutational multivariate ANOVA; KW, Kruskal–Wallis rank sum test