Figure 3.
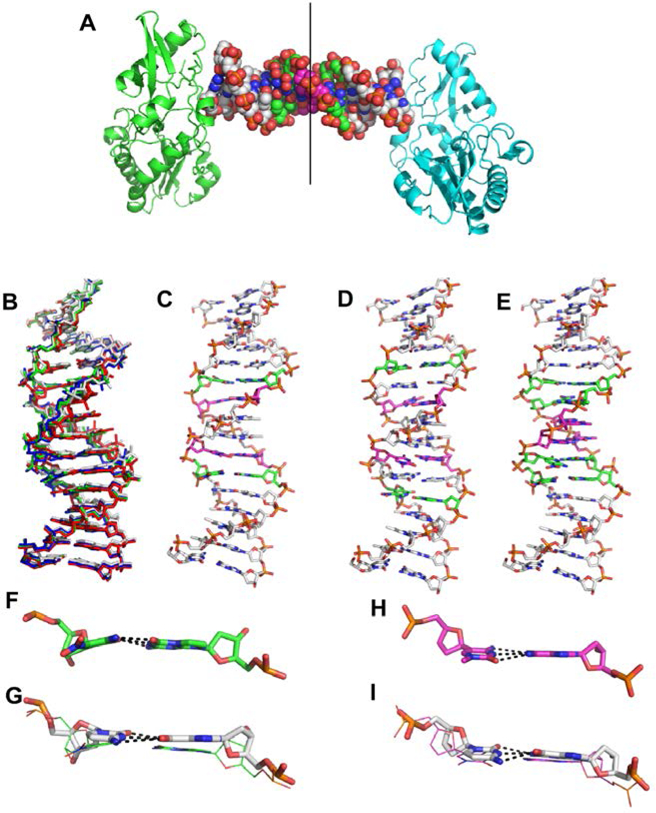
Crystal structures of PB, PC, and PP hachimoji DNA. (A) The host-guest complex with two N-terminal fragments from Moloney murine leukemia virus reverse transcriptase in green and cyan bound to a 16mer PP hachimoji DNA; in the duplex sphere model, Z:P pairs are green, S:B pairs magenta. The asymmetric unit includes one protein molecule and half of the 16mer DNA as indicated by the line. (B) Hachimoji DNA structures PB (green), PC (red), PP (blue) are superimposed with GC DNA (gray). (C) Structure of hachimoji DNA with self- complementary duplex CTTATPBTASZATAAG (“PB”). (D) Structure of hachimoji DNA with self-complementary duplex 5’-CTTAPCBTASGZTAAG, (“PC”). (E) Structure of hachimoji DNA with self-complementary duplex with six consecutive non-standard components 5’-CTTATPPSBZZATAAG (PP). DNA structures are shown as stick models with P:Z pairs (C, green), B:S pairs (C, magenta), and natural pairs (C, gray). Examples of largest differences in detailed structures: (F) The Z:P pair (from the PB structure) is more buckled than corresponding G:C pair in (G). (H) The S:B pair (from the PB structure) exhibits a propeller angle similar to that in the corresponding G:C pair shown in (I).
