Fig. 4.
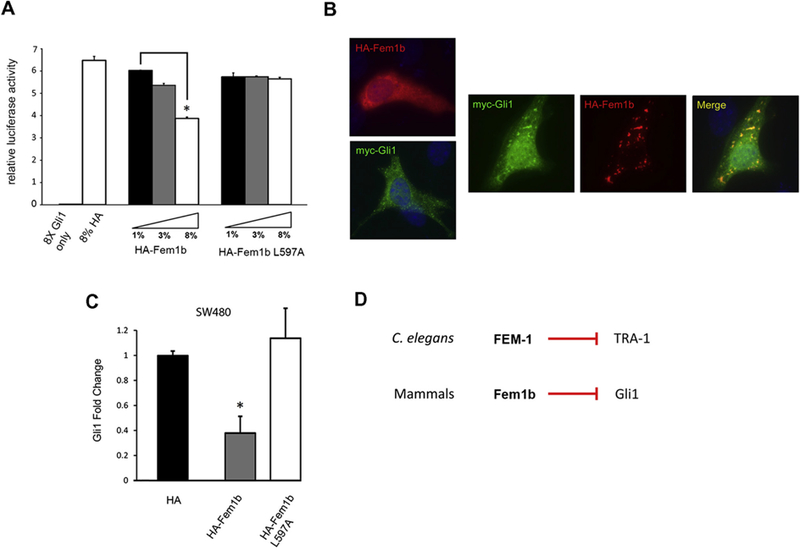
Fem1b suppresses Gli1 transcriptional activity. (A) Luciferase activity of NIH-3T3 cells transfected with a Gli1 luciferase reporter and the indicated constructs was measured (data is a representative experiment with error bars expressing the average relative luciferase activity ± the standard deviation (S.D.) from three repeated measurements); ∗p < 0.01. (B) Ectopically expressed Fem1b and Gli1 co-localize. Left: NIH-3T3 cells were transfected with either HA-Fem1b or Myc-Gli1 for 24 h and fixed and immunostained for HA (red) or Myc (green). Right: NIH-3T3 cells were co-transfected with HA-Fem1b and Myc-Gli1 and fixed and immunostained for HA (red) and Myc (green). Co-localized proteins appear yellow. Nuclei were stained by 4’,6-diamino-2-phenylindole (DAPI) (blue). (C) Transient expression of Fem1b reduces the level of Gli1 message. The qPCR was performed on total RNA isolated from SW-480 cells transfected with the indicated constructs after 48 h. Gli1 message was determined relative to GAPDH message levels. Data are normalized to Gli1 message in empty vector transfected cells (error bars expressed as mean fold induction ± standard deviation (S.D.); ∗p < 0.05) and reflect three independent experiments and (D) in the C. elegans sex determination pathway FEM-1 negatively regulates the zinc finger transcription factor TRA-1 (above), similarly in the mammalian Hedgehog signaling pathway Fem1b negatively regulates the homologous zinc finger transcription factor Gli1 (below).
