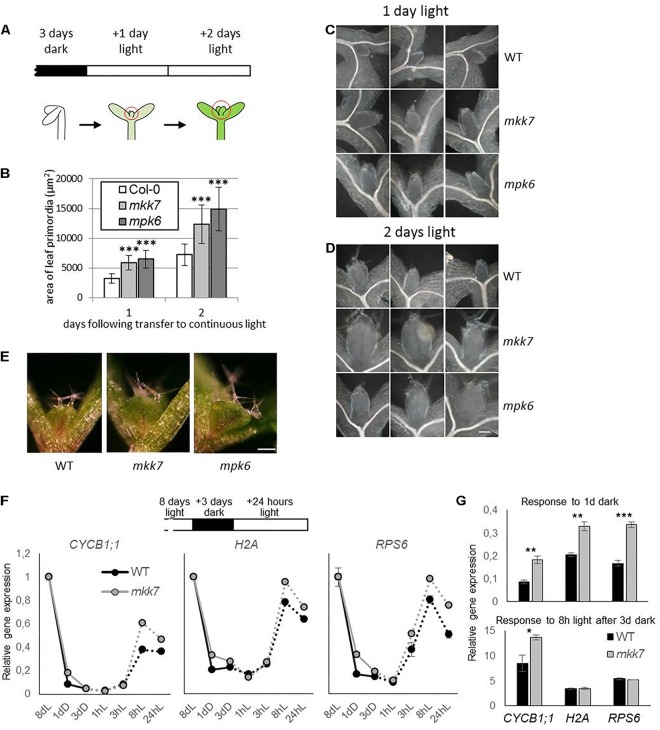
FIGURE 1.
Utilization of seedling de-etiolation as a synchronized developmental model. (A) De-etiolation experimental setup. Seeds were germinated for 3 days in darkness then transferred to continuous light. Area of developing leaf primordia were quantified by images of microscopic preparations of seedlings made 1 or 2 days following transfer. (B) Area of developing Col-0, mpk6, and mkk7 leaf primordia at 1 and 2 days in continuous light, following 3 days growth in dark. About 30 seedlings were used for each sample. P-values: 1.32E-09–2.29E-14, indicated by three asterisks. The error bars represent standard deviation. (C,D) Representative images of Col-0, mkk7, and mpk6 leaf primordia at one (C) and two (D) days following exposure to light. Scale bar: 50 μm. (E) Col-0, mkk7, and mpk6 seedlings germinated in darkness for 3 days then exposed to continuous light for further 3 days. Scale bar: 250 μm. The images are representative of at least 12 seedlings. (F,G) Relative gene expression of signature genes during dark arrest and subsequent light exposure in emerging leaf primordia. Wild-type seedlings were gown in light on sucrose-containing plates for 8 days, transferred to dark and returned to light after three subsequent days. Two hundred seedlings were harvested for each biological replicate at the corresponding times and had the primordia of leaves 1 and 2 dissected. Gene expression was monitored by quantitative real-time PCR and shown as the entire time course (F) and for specific time points relative to that at the onset of dark treatment or at the onset of the light treatment (G). Error bars indicate standard error of the mean (between biological replicates). Asterisks indicate level of significance: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.