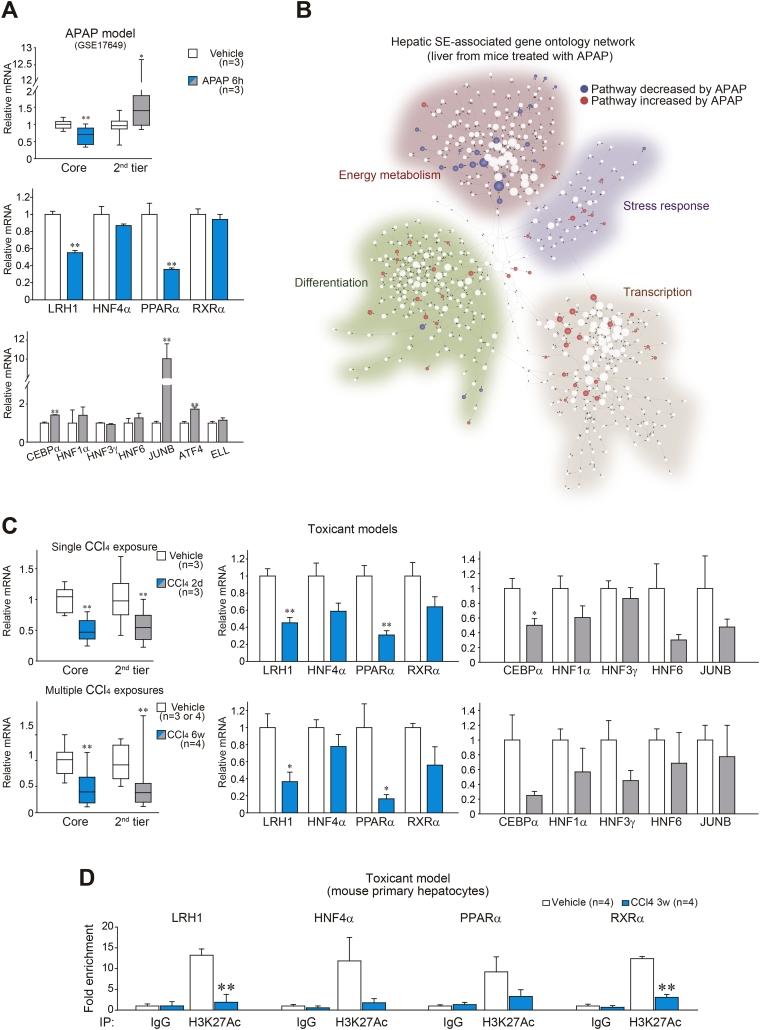
Fig. 3.
Loss of core TFs in mouse liver disease models.
(A) TF transcript levels in the liver of mice treated with APAP. Data were extracted from GSE17649 (Affymetrix global scaling normalization). Gene expression changes in liver were calculated relative to vehicle-treated group.
(B) Hepatic super-enhancer-associated GO network displaying RNA-seq expression data from the liver of mice treated with APAP. Decreased or increased pathways were depicted as a blue or red node, respectively. RNA-seq data were generated using the mouse liver tissues obtained six h after APAP treatment. RNA-seq data are deposited in the GEO under accession number GSE104302.
(C) TF transcript levels in the injured liver of mice. The mice were sacrificed two days after a single injection of CCl4, or multiple injections of CCl4 for six weeks.
(D) ChIP assays. Crosslinked protein–DNA complexes were immunoprecipitated using anti-H3K27Ac antibody or preimmune-IgG (negative control) in primary hepatocytes isolated from mice treated with vehicle or CCl4 for three weeks. qPCR assays were done to quantify DNAs in the immunoprecipitates using specific primers for each super-enhancer region.
Data information: For upper panel of A and left of C, data were shown as box and whisker plot (significantly different as compared with vehicle-treated control, *P < .05; **P < .01). Box, IQR; whiskers, 5–95 percentiles; and horizontal line within box, median. For lower panels of A and middle and right of C, data represent the means ± SEM (significantly different as compared with vehicle-treated controls, *P < .05; **P < .01). Group sizes (n) are denoted in each figure. For D, data represent the means ± SEM (n = 4 each, significantly different as compared to vehicle control, **P < .01). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)