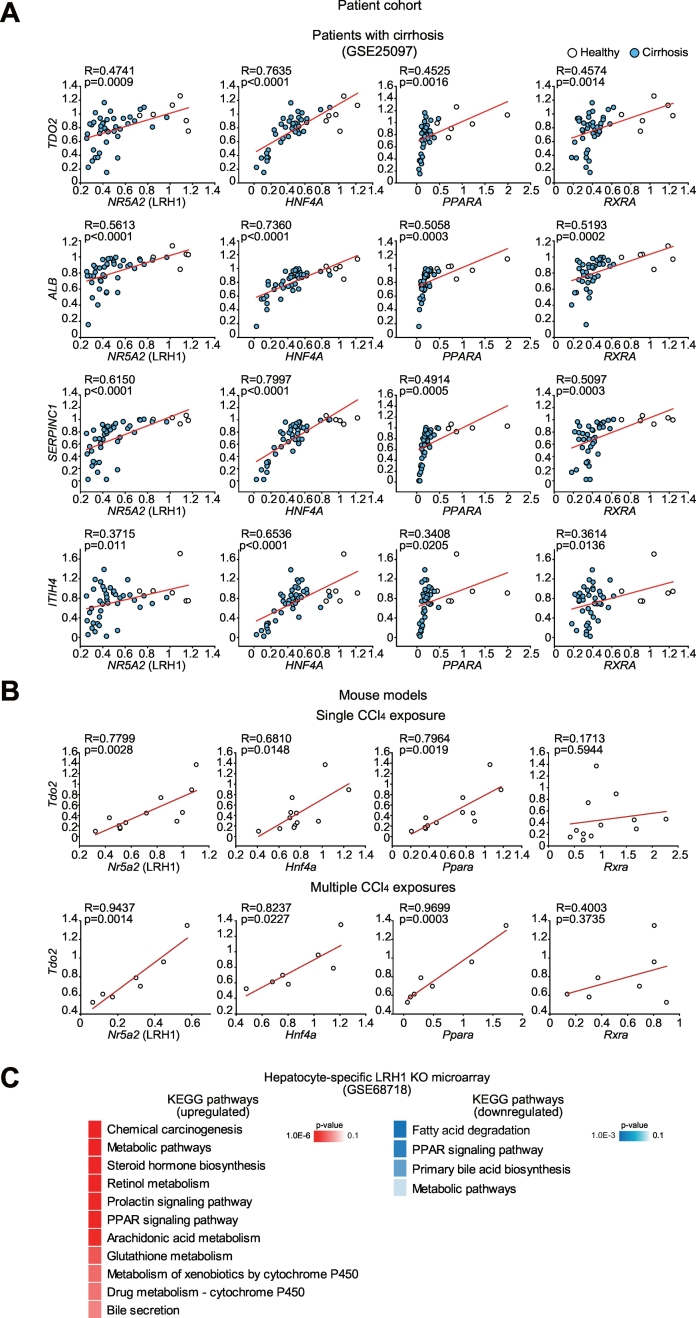
Fig. 4.
Correlation between identified core TF and each gene transcript levels.
(A) Correlation analyses in a large cohort of cirrhosis patients (GSE25097) (N = 46). Pearson's r correlation coefficients with corresponding P-values for co-variation between each core TF mRNA levels (x axis) and hepatocyte identity gene transcripts (y axis) show robust correlations.
(B) Pearson correlation analyses in mice treated as in Fig. 3C. For single and multiple CCl4 treatment models, group sizes (N) are 12 and 7, respectively.
(C) Results of KEGG pathway analysis of the up- or down-regulated genes after hepatocyte-specific deletion of LRH1 (GSE68718). Enriched signaling pathways of each gene cluster were analyzed using DAVID.