Fig. 6.
LRH1-dependent recovery of hepatocyte identity genes.
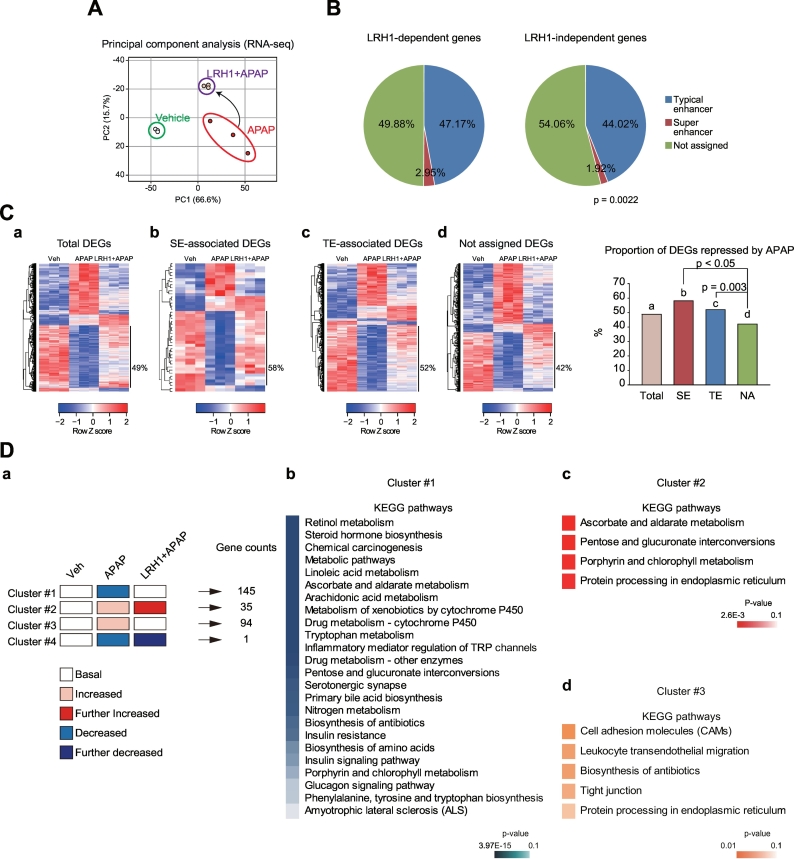
(A) Principal component analysis of RNA-seq data. RNA-seq data was generated using liver of mice treated with APAP alone or APAP+LRH1 overexpression vector as in Fig. 5A. RNA-seq data are deposited in the GEO under accession number GSE104302.
(B) Pie graphs showing the enhancer composition. Genes having P-values lower than 0.05 between APAP and LRH1 + APAP groups were defined as LRH1-dependent genes.
(C) Heatmaps and hierarchical correlation analyses of differentially expressed genes (DEGs). DEGs were selected as the genes with independent t-test (P-values < .05 with a fold-change of >1.5). The DEGs were hierarchically clustered and presented as heatmaps according to the row Z score. Super-enhancer- or typical enhancer (SE or TE)-associated DEGs represent significantly altered genes in the APAP group as compared to the vehicle group among the SE-associated or TE-associated genes. Not assigned DEGs are DEGs which are assigned neither to SE nor TE. Heatmaps of total DEGs (a), SE-associated DEGs (b), TE-associated DEGs (c), and not assigned DEGs (d) are presented in left. The proportions of the gene clusters depicted in the heatmaps (left) were shown as a graph (right). (D) Results of KEGG pathway analysis of the clustered DEGs. A schematic description of the gene clusters (a). Enriched signaling pathways of each gene cluster were analyzed using DAVID (b-d).