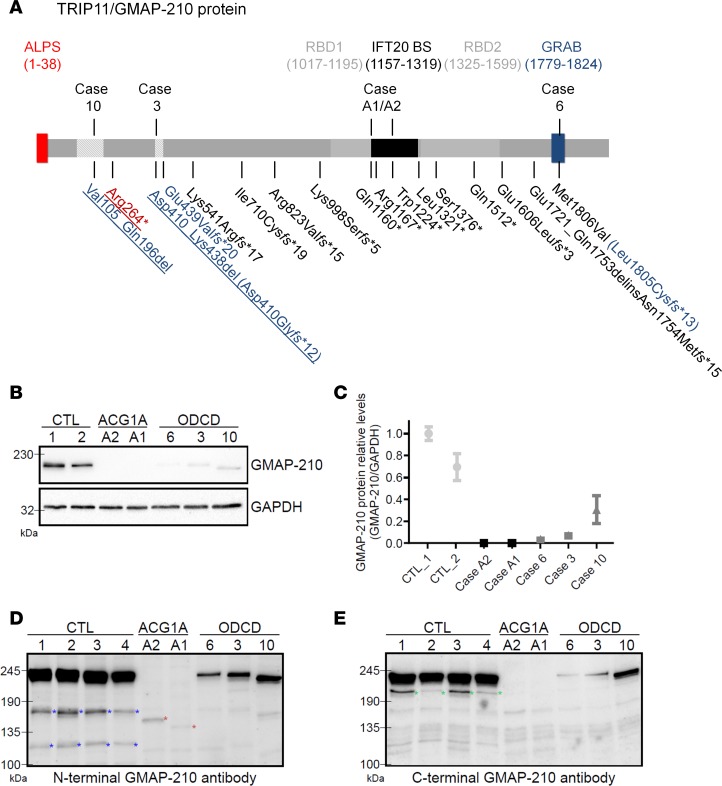
Figure 4. GMAP-210 protein analysis in achondrogenesis 1A (ACG1A) and odontochondrodysplasia (ODCD) patient–derived primary cells.
(A) Schematic of the 1,979 amino acid GMAP-210 protein. The backbone of coiled-coil domains is shown in gray; the amphipathic lipid sensor (ALPS) in red, which participates in vesicle tethering with an overlapping second motif (not shown); Rab-binding domains 1 and 2 (RBD1 and RBD2) in light gray, which mediate RAB2 binding; and the GRIP-related Arf-binding (GRAB) domain in blue, which mediates membrane anchoring of GMAP. The IFT20 binding site (IFT20 BS, in black) is necessary for Golgi targeting of IFT20. Mutations identified in 5 ACG1A and 10 ODCD cases are indicated below; those amenable to functional analysis are indicated above. Amino acids deleted by in-frame exon skipping are hatched; nonsense and frameshift mutations are depicted in black, splice mutations in blue, and mutations shared by ACG1A and ODCD in red; recurrent mutations are underlined. (B) GMAP proteins in whole-cell protein lysates of patients and controls; GAPDH staining demonstrates total protein loading. The mean of triplicate quantitative blot signal analyses is represented in C; error bars indicate ± SEM. (D–E) GMAP-210 in whole-cell protein lysates of patients and controls using more sensitive enhanced chemiluminescence (ECL) substrate to analyze low-abundant GMAP protein variants with polyclonal antibodies directed against the N-terminus and the C-terminus. (D) Note the ~3 kDa lower protein variant in case 3 (consistent with the loss of exon 9) and the ~10 kDa lower protein variant in case 10 (consistent with the loss of exon 4). Red asterisks mark specific signals in the ACG1A cases (consistent with biallelic stop mutations and a predicted protein size of about 142 kDa in case A2 and 134 kDa in case A1). Additional bands for GMAP-210 marked with blue asterisks in CTL cells may represent shorter variants or proteolytic cleavage products. (E) Green asterisks mark the putative GMAP-190 protein corresponding to TRIP11-ΔEx9short.