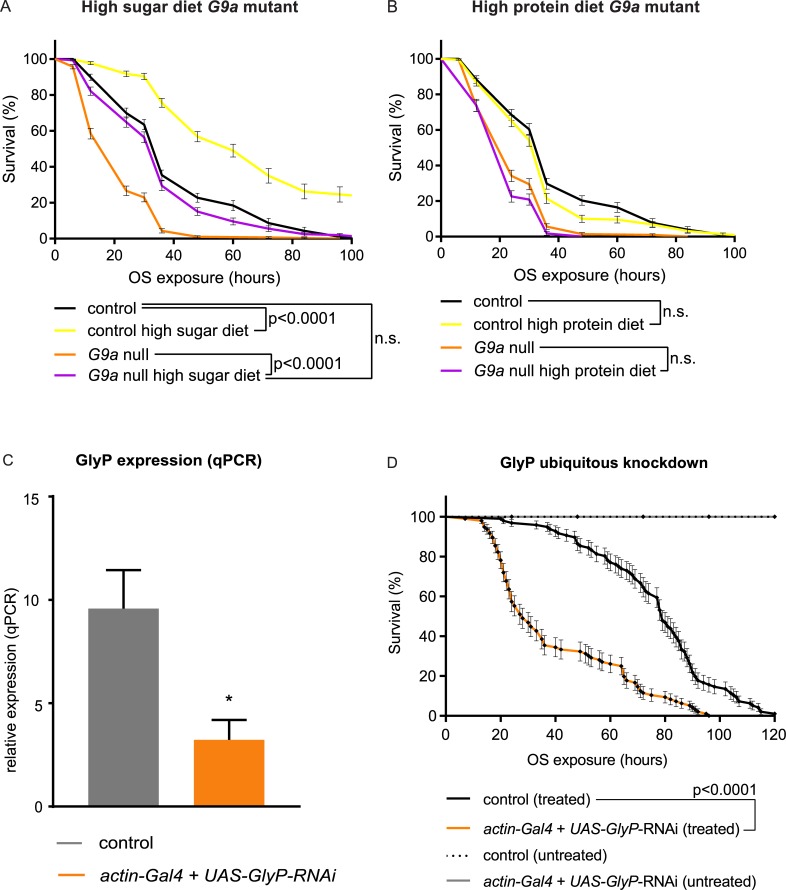
Fig 8. OS-mediated survival defects of G9a mutants can be rescued by a high-sugar diet and are recapitulated by inhibition of glycogen breakdown.
(A, B) Survival curves of G9a mutants and controls fed or not with a high-sugar (A) or high-protein (B) diet during OS exposure. (C) GlyP relative expression in ubiquitous GlyP-knockdown flies (actin-Gal4 + UAS-GlyP-RNAi) and controls (actin-Gal4), as determined by qPCR. To generate these flies, the ubiquitously expressed actin driver was combined with a UAS-GlyP RNAi construct (actin-Gal4 + UAS-GlyP-RNAi) to knock down GlyP expression or crossed to the isogenic background of the RNAi construct to generate the isogenic control (actin-Gal4). (D) Survival curves of GlyP ubiquitous knockdown flies and controls (actin-Gal4, treated) upon paraquat-induced OS (treated) or untreated. Genotypes are as in (C). Median survival time actin-Gal4 + UAS-GlyP-RNAi (treated): 28 h, n = 96 versus actin-Gal4 (treated) 79 h, n = 96; p < 0.0001. Survival curves showing percent survival and SE over time were plotted using Graphpad, and p-values were obtained using the Gehan-Breslow-Wilcoxon test. The numerical data depicted in this figure can be found in S5 Data. GlyP, glycogen phosphorylase; n.s., not significant; OS, oxidative stress; qPCR, quantitative PCR; RNAi, RNA interference; SE, standard error.