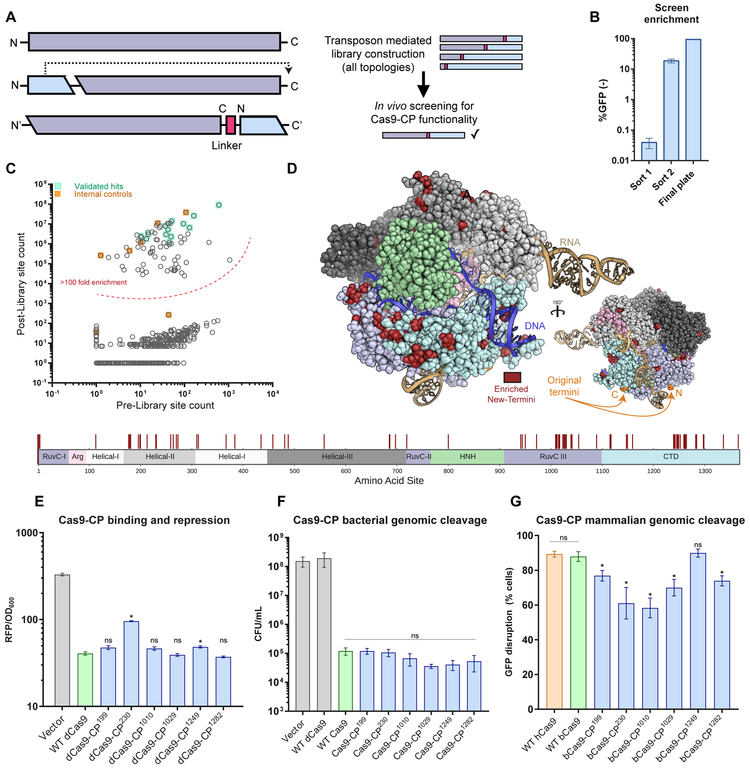
Figure 1. An unbiased Cas9 library screen identifies active circularly permuted Cas9 proteins.
(A) Overview of circular permutation and library generation for Cas9.
(B) Enrichment values of the unbiased screen as determined by flow cytometry and colony forming units. Error bars indicate standard deviation in all panels.
(C) Deep sequencing read averages for the Pre- and Post- Cas9-CP library members, demonstrating a strong clustering of highly enriched library members with internal (within 4 AA of the N- and C-termini) and empirically validated controls. The dotted line highlights an approximate boundary that represents >100-fold enrichment in the screen.
(D) Model of new Cas9-CP termini (in red) based on PDB ID: 5F9R with domains colored according to the sequence bar (below). New termini are mapped onto the AA sequence bar.
(E) Endpoint values for dCas9-CP 12 hr E. coli CRISPRi DNA binding and RFP repression system compared with WT dCas9 and a protein expression vector control in triplicate (error bars are s.d., * = p<0.05, ns = not significant, t-test).
(F) CFU/mL readings in an E. coli genomic cleavage assay read out by cell death compared with a protein expression vector control, WT dCas9 and WT Cas9 (n = 3, error bars are s.d., * = p<0.05, ns = not significant, t-test).
(G) Cleavage efficiency of a genomic reporter in mammalian cells in triplicate (described in Figures S2B and S2C), observed via indel formation and GFP reporter disruption. hCas9 is human codon optimized Cas9; bCas9 indicates bacterial codon-based Cas9 constructs (error bars are s.d., * = p<0.05, ns = not significant, t-test).