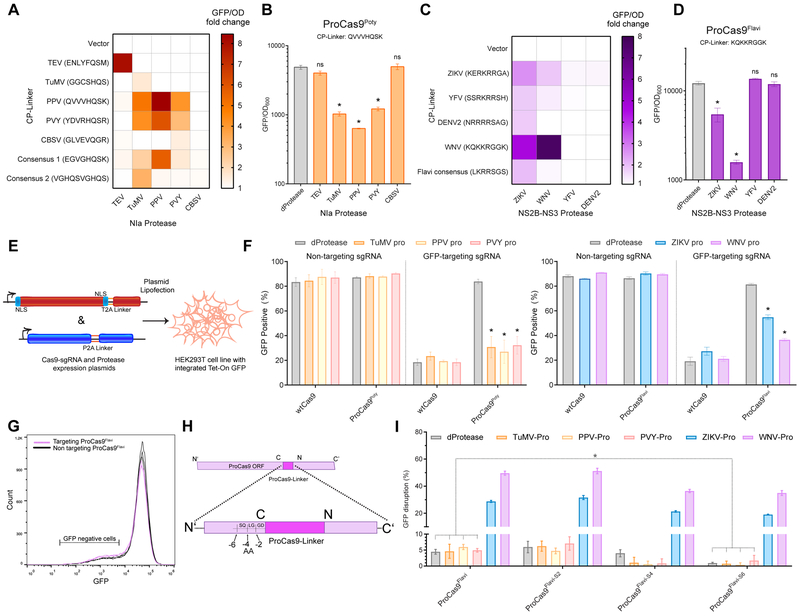
Figure 3. Generation of ProCas9s for sensing and responding to Potyvirus and Flavivirus proteases.
(A) Heat map depicting the fold activation of a suite of ProCas9 CP linkers for Potyviral NIa proteases. Data is normalized to a non-active protein expression control (dTEV) in an E. coli based CRISPRi GFP repression assay. Darker coloration indicates greater activity (n = 2).
(B) Endpoint analysis of the E. coli CRISPRi assay utilizing the linker derived from Plum Pox Virus (PPV) comparing the response to distinct NIa proteases and a dead protease (n = 3, error bars are s.d., * = p<0.05, ns = not significant, t-test compared to dProtease).
(C) Heat map depicting the fold activation of a suite of ProCas9 CP linkers for Flavivirus NS2B-NS3 proteases, normalized to a non-active protein expression control (dTEV) in an E. coli based CRISPRi GFP repression assay. Darker coloration indicates greater activity (n = 2).
(D) Endpoint analysis of the E. coli CRISPRi assay utilizing the linker derived from West Nile Virus (WNV) showing the response to distinct NS2B-NS3 proteases and a dead protease (n = 3, error bars are s.d., * = p<0.05, ns = not significant, t-test compared to dProtease).
(E) Schematic of the constructs used for the transient transfection and testing in HEK293T cells.
(F) Mammalian GFP disruption assay (Figures S2A-C). HEK293T-based reporter cells were transfected with the indicated sgRNAs, WT Cas9 or a ProCas9 variant, and the respective proteases Reduction in GFP positive cells indicates genome cleavage by a Cas9 construct (n = 3, error bars are s.d., * = p<0.05, t-test compared to dProtease).
(G) Flow cytometry plots from (F) with overlay of GFP-targeting (pink) vs. non-targeting (black) ProCas9Flavi systems, demonstrating a small degree of background activity.
(H) Truncation of ProCas9 AA linker sequence to prevent leakiness.
(I) Leakiness and orthogonality of the original and shortened ProCas9Flavi constructs. Displayed as percent GFP disrupted via normalization to the non-targeting guide for each construct-protease pairing. In addition to the deactivated protease (dProtease) control, the active Potyvirus NIa proteases were used to assess orthogonality (n = 3, error bars are s.d., * = p<0.05, ns = not significant, t-test).