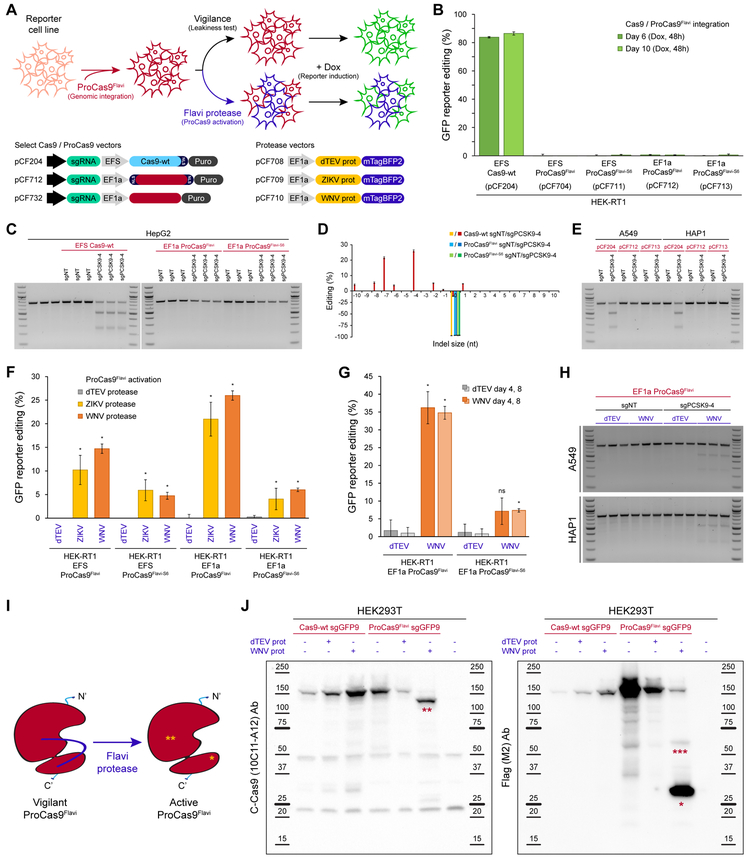
Figure 4. ProCas9 stably integrated into mammalian genomes can sense and respond to Flavivirus proteases.
(A) Genomic integration and testing of Flavivirus protease-sensitive ProCas9s. HEK-RT1 genome editing reporter cells are stably transduced with various ProCas9 lentiviral vectors, followed by puromycin selection of ProCas9 cell lines. These cell lines are then either tested for leaky ProCas9 activity in the absence of a stimulus, or stably transduced with a vector expressing the indicated proteases, followed by assessment of genome editing using the GFP reporter.
(B) Leakiness assessment of ProCas9 variants expressed from either the EFS or EF1a promoter. HEK-RT1 reporter cells were stably transduced with the indicated ProCas9 variants or Cas9-wt. Genome editing activity was quantified at the indicated days post-transduction. Error bars indicate the standard deviation of triplicates.
(C) Leakiness assessment at the endogenous PCSK9 locus. HepG2 cells stably transduced with the indicated sgRNAs and ProCas9 variants or Cas9-wt. Cells were selected on puromycin and harvested at day 8 post-transduction for T7E1 analysis.
(D) Mutational patterns and editing efficiency at the PCSK9 locus of samples shown in (C). Indels were quantified using TIDE. For clarity, the fraction of non-edited cells is represented as negative percentages.
(E) ProCas9 leakiness quantification, as in (C), in A549 and HAP1 cells. Cells were selected on puromycin and harvested at day 7 post-transduction for T7E1 analysis.
(F) Quantification of Flavivirus ProCas9 activation in response to various control (dTEV, pCF708) or Flavivirus (ZIKV, pCF709; WNV, pCF710) proteases. ProCas9 reporter cell lines were stably transduced with the indicated protease vectors. At day 3 post-transduction, cells were treated with doxycycline to induce GFP reporter expression. Error bars indicate the standard deviation of triplicates. Significance was assessed by comparing each sample to its respective dTEV control (unpaired, two-tailed t-test, n = 3, * = p<0.05, ns = not significant).
(G) Genome editing activity in Flavivirus ProCas9 reporter cell lines, as in (F), at day 4 or 8 post-transduction.
(H) Protease-sensitive editing at the endogenous PCSK9 locus. T7E1 assay of A549 and HAP1 Flavivirus ProCas9 cell lines (sgNT, sgPCSK9-4) stably transduced with the indicated mTagBFP2-tagged viral proteases. At day 4 post-transduction, mTagBFP2-positive cells were sorted and harvested for T7E1 analysis.
(I) ProCas9Flavi activation by Flavivirus (Flavi) proteases. *, small subunit of the activated ProCas9Flavi (29 kDa). **, large subunit of the activated ProCas9Flavi (137 kDa).
(J) Immunoblotting for Cas9 in HEK293T co-transfected with plasmids expressing Cas9-wt or ProCas9Flavi and dTEV or WNV proteases. The C-Cas9 (clone 10C11-A12) antibody recognize the large subunit of the activated ProCas9Flavi (**, 137 kDa). The Flag-tag (clone M2) antibody recognizes the small subunit of the activated ProCas9Flavi (*, 29 kDa). ***, likely small-subunit-ProCas9Flavi-T2A-mCherry (55 kDa). Protein ladders indicate reference molecular weight markers.