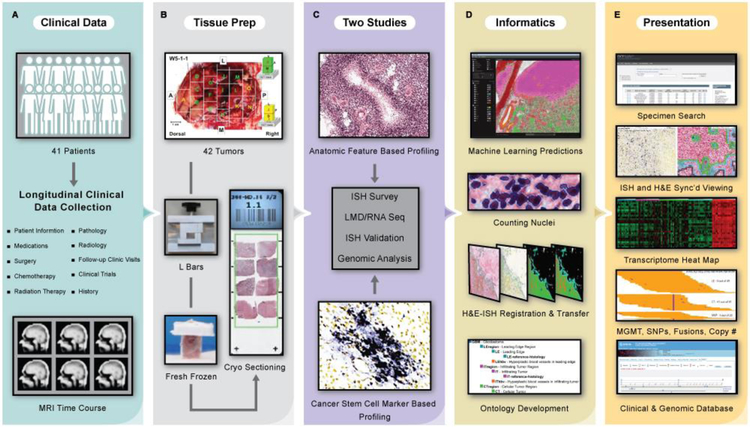
Fig. 1. Data generation, analysis, and presentation pipeline for the Ivy Glioblastoma Atlas Project.
(A) Clinical data were collected for the Ivy cohort of 41 patients. (B) Tissue preparation required en bloc resection and formation of tissue blocks with custom L bars. (C) Two studies, Anatomic Feature Based Profiling and Cancer Stem Cell Marker Based Profiling, provided a framework for the ISH surveys, LMD/RNA-Seq experiments, and ISH validations. (D) Informatics included image registration, ontology development, and anatomic feature prediction based on a novel machine learning (ML) analysis of histological data. Search tools support queries of the data set by tumor, tumor block, and gene expression filtered by anatomic feature, molecular subtype, and clinical information. Searchable manual labels delineating the laser microdissections for 270 RNA-Seq samples from the two studies overlay the histology images. The atlas is equipped with image viewers that resolve the histology at 0.5μm/pixel, a transcriptome browser, an application programming interface, and help documentation. The database has detailed longitudinal clinical information and MRI time courses (table S1). (E) This free resource is made available as part of the Ivy Glioblastoma Atlas Project (Ivy GAP). (http://glioblastoma.alleninstitute.org/) via the Allen Institute data portal (http://www.brain-map.org), the Ivy GAP Clinical and Genomic Database (http://ivygap.org/) via the Swedish Neuroscience Institute (http://www.swedish.org/services/neuroscience-institute), and The Cancer Imaging Archive (https://wiki.cancerimagingarchive.net).