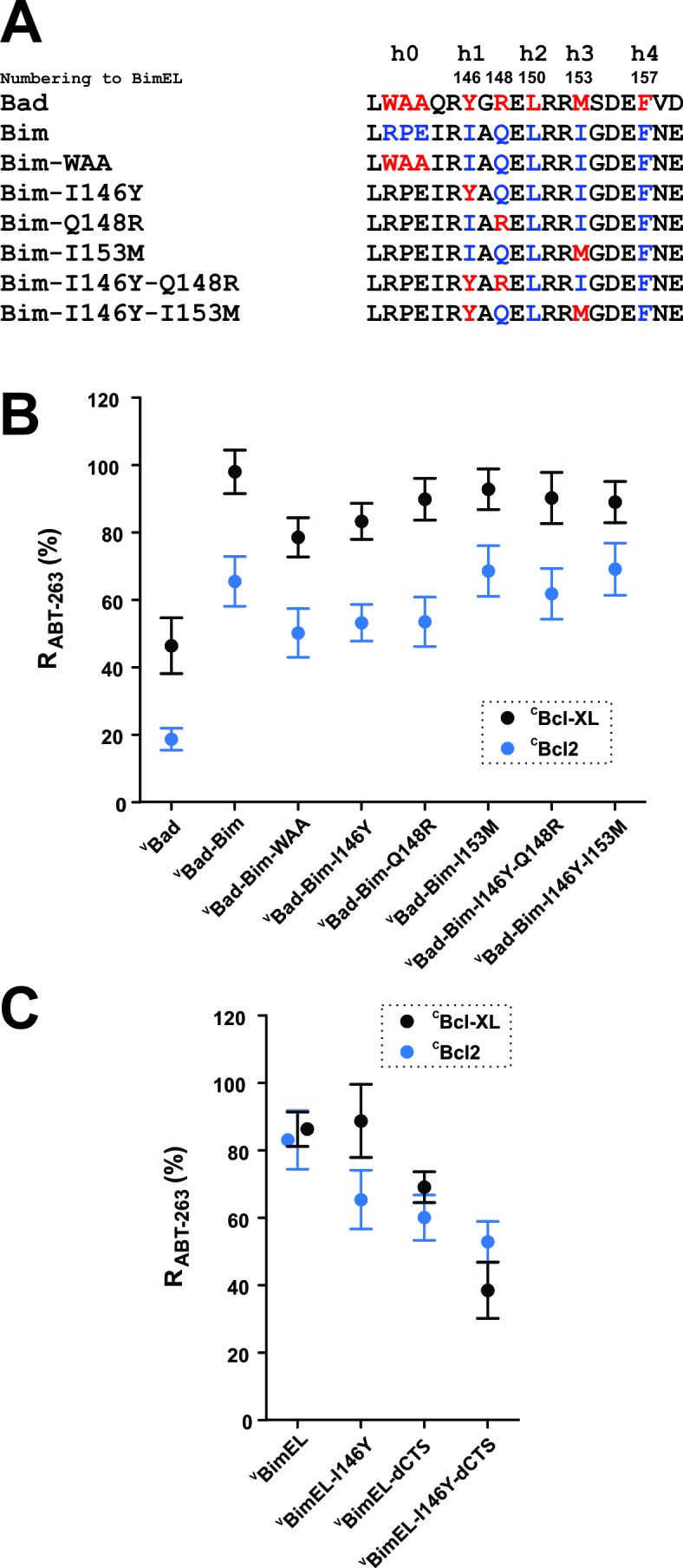
Figure 6. h0 and h1 residues in the Bim BH3 contribute to the resistance of Bcl-XL:Bim complexes to ABT-263.
(A) Sequence alignment of the BH3 regions of Bad and Bim and the sequences of VBad-Bim mutants, residues from Bad (red), Bim (blue). (B) Identification of conserved hydrophobic residues in the Bim BH3 that contributed to RABT-263 for CBcl-XL:VBad-Bim (black) and CBcl-2:VBad-Bim (blue) complexes. The RABT-263 values of CBcl-XL:VBad-Bim and CBcl-2:VBad-Bim complexes from Figure 4b and Table 1 were included to facilitate direct comparison. (C) The BH3 h1 residue I146 contributes to Bim binding to Bcl-2 and Bcl-XL. Substitution of I146 with the corresponding residue from Bad (BimEL-I146Y) decreased RABT-263 for CBcl-XL: VBimEL/VBimEL-dCTS (black) and CBcl-2:VBimEL/VBimEL-dCTS (blue) complexes. Data are mean ±95% confidence intervals calculated from FLIM-FRET binding curves shown in Figure 6—figure supplement 1.