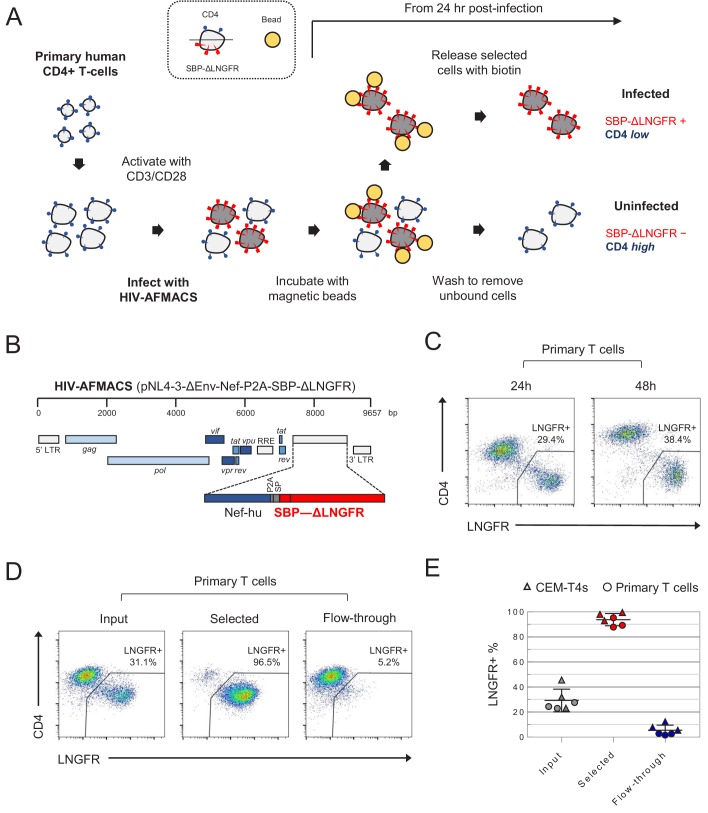
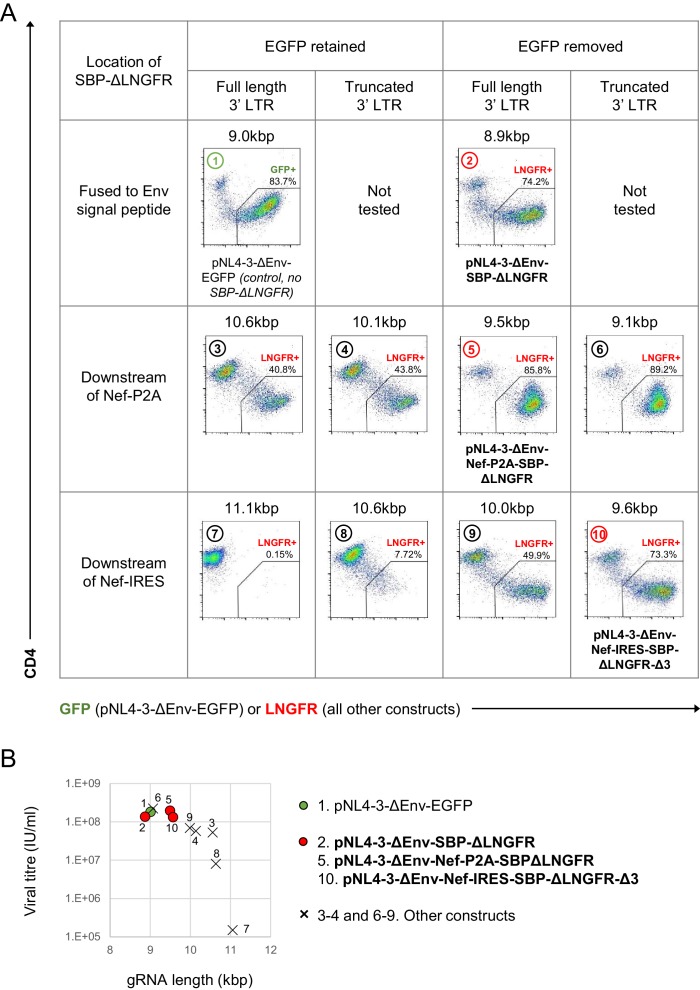
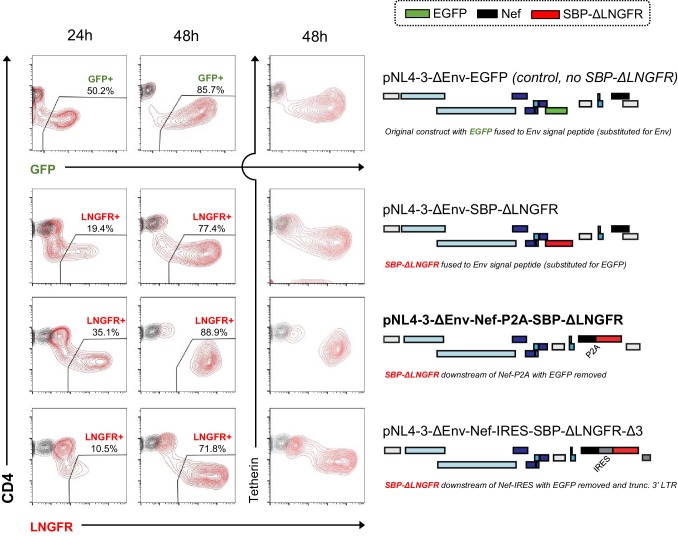
Figure 1. Antibody-free magnetic selection of HIV-infected primary T cells.
(A) Workflow for AFMACS-based magnetic selection of HIV-infected primary T cells. (B) Schematic of HIV-AFMACS provirus (pNL4-3-ΔEnv-Nef-P2A-SBP-ΔLNGFR). For simplicity, reading frames are drawn to match the HXB2 HIV-1 reference genome. Length is indicated in base pairs (bp). The complete sequence is available in Supplementary file 1. Nef-hu, codon-optimised Nef; RRE, Rev response element; SP, signal peptide. (C) Expression of cell surface SBP-∆LNGFR and CD4 on primary T cells 24 or 48 hr post-infection with HIV-AFMACS. Cells were stained with anti-LNGFR and anti-CD4 antibodies at the indicated time points and analysed by flow cytometry. (D–E) Magnetic sorting of HIV-infected (red, LNGFR+, CD4 low) and uninfected (blue, LNGFR-, CD4 high) cells. Cells were separated using AFMACS 48 hr post-infection with HIV-AFMACS and analysed as in (C). Representative (D) and summary (E) data from six independent experiments in CEM-T4s (triangles) and primary T cells (circles) are shown, with means and 95% confidence intervals (CIs).