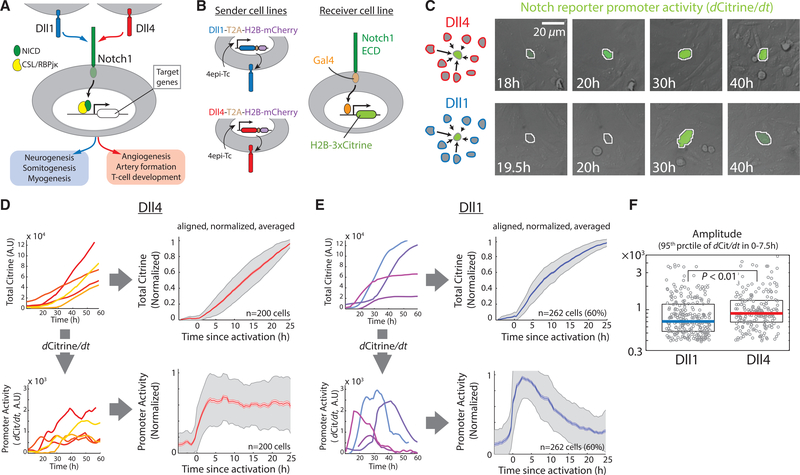
Figure 1. Dll1 and Dll4 Activate Notch1 with Pulsatile and Sustained Dynamics, Respectively.
(A) Both Dll1 (blue) and Dll4 (red) activate the Notch1 receptor (green) to induce proteolytic release of the Notch intracellular domain (NICD), but are used in different biological contexts (blue and red boxes, bottom). The released NICD translocates to the nucleus and, in complex with CSL/RBPjκ (yellow), activates Notch target genes (white).
(B) Left: Engineered CHO-K1 “sender” cell lines contain stably integrated constructs expressing Dll1 (blue) or Dll4 (red), each with a co-translational (T2A, brown) H2B-mCh readout (purple), from a 4epi-Tetracycline (4epi-Tc) inducible promoter. Right: “Receiver” cells stably express a chimeric receptor combining the Notch1 extracellular domain (Notch1ECD) with a Gal4 transcription factor (orange), which can activate a stably integrated fluorescent H2B-3xCitrine reporter gene (chartreuse).
(C) Left (schematics): A minority of receiver cells (green) are co-cultured with an excess of either Dll1 (blue) or Dll4 (red) sender cells. Right: Filmstrips showing representative sustained (top, Dll4 senders) or pulsatile (bottom, Dll1 senders) response of a single receiver cell (center, automatically segmented nucleus outlined in white). Grey channel shows DIC images of cells, while the rate of increase in Citrine fluorescence, scaled to 25%–75% of its total range, is indicated using green pseudo-coloring. See also Movies S1 and S2.
(D) Left: Representative traces showing total nuclear Citrine fluorescence levels (top) or corresponding derivatives of the total Citrine (dCitrine/dt), i.e., promoter activity (bottom), in individual receiver cells activated by Dll4. Right: Average values of total fluorescence (top) and promoter activity (bottom) in receiver cells activated by Dll4. Solid traces represent medians, lighter shades indicate SEM, and gray shading indicates SD. n, number of traces included in the alignment. See STAR Methods for alignment and normalization procedure.
(E) Left: Corresponding plots (as in D) showing total nuclear Citrine fluorescence levels (top) and promoter activity (bottom) in individual receiver cells in co-culture with Dll1. Right: Average values of total fluorescence (top) and promoter activity (bottom) in receiver cells activated by Dll1. The percentage value (60%) in the plots on right indicates the fraction of receiver traces included in the alignment (STAR Methods, see also Figure S1F).
(F) 95th percentile of (absolute, non-normalized) promoter activity values between 0 and 7.5 hr (after alignment) in the traces included in (D) and (E). This time window is chosen to simultaneously estimate the promoter activity at the peak of Dll1 pulses and at steady-state levels of Dll4 signaling. Solid horizontal lines represent medians, while the boxes delineate 25th–75th percentile values. p value calculated by two-sided Kolmogorov-Smirnov (K-S) test.
See also Figures S1 and S2.