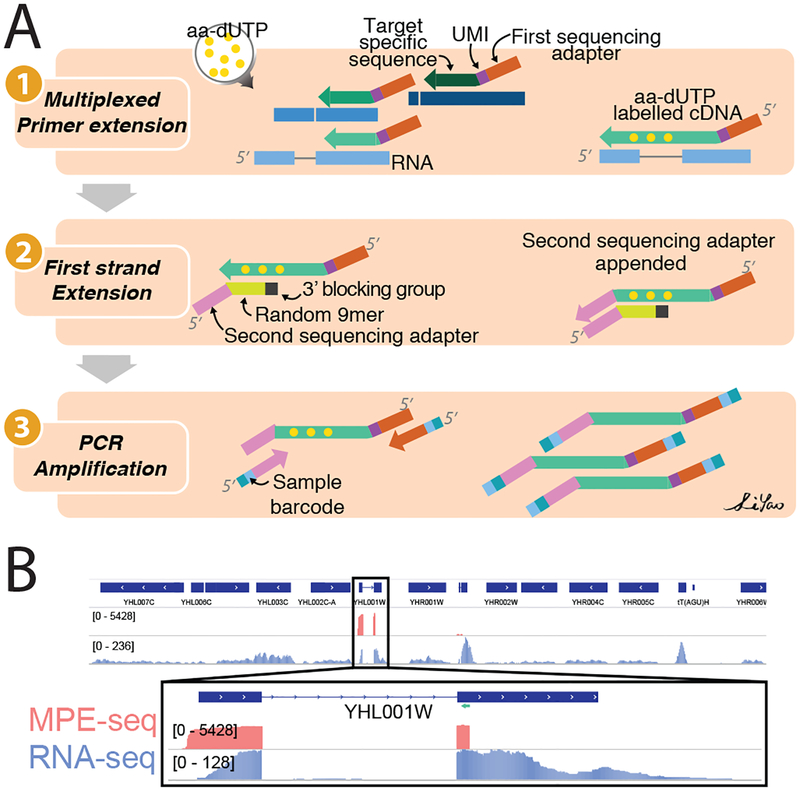
Figure1: MPE-seq uses complex pools of reverse transcription primers to target sequencing to regions of interest.
(A) Outline of MPE-seq protocol. (1) Hundreds to thousands of primers are pooled and extended by reverse transcription at targeted regions of interest. The incorporation of amino-allyl dUTP (aa-dUTP) nucleotides during reverse transcription enables purification of cDNA between subsequent steps. (2) The second sequencing adapter is appended to the 3’ terminus of the cDNA using an oligonucleotide template and Klenow enzyme. (3) Libraries are PCR amplified and sequenced.
(B) Genome browser screenshot of a targeted region in MPE-seq (pink) and conventional RNA-seq (purple). The location of a targeting primer is shown as a green arrow.