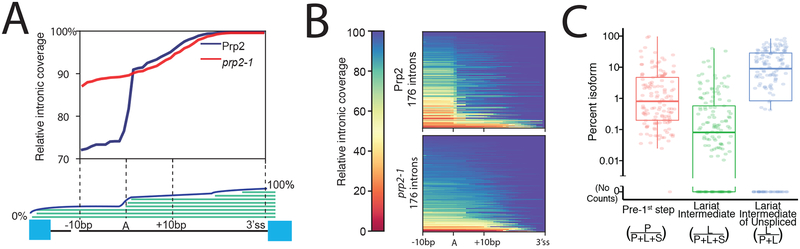
Figure3: MPE-seq enables genome-wide profiling of lariat intermediates.
(A) Meta-intron coverage plot surrounding predicted branchpoints in a wild-type (Prp2) and step1 splicing mutant strain (prp2–1). The region between the +10 position downstream of the annotated branchpoint and the 3′ splice site (3′ss) was re-scaled for each intron.
(B) Heatmap plots showing the relative coverage at each intron for which lariat intermediate reads were detected.
(C) Estimates of the relative abundance of each isoform for each targeted intron for which reads were detected. Horizontal lines in boxplots represent the 25th, 50th, and 75th percentiles. Whiskers end at the 0th and 100th percentiles. n=141 introns for which we attempted lariat quantification and found at least one spliced read.