Figure 2.
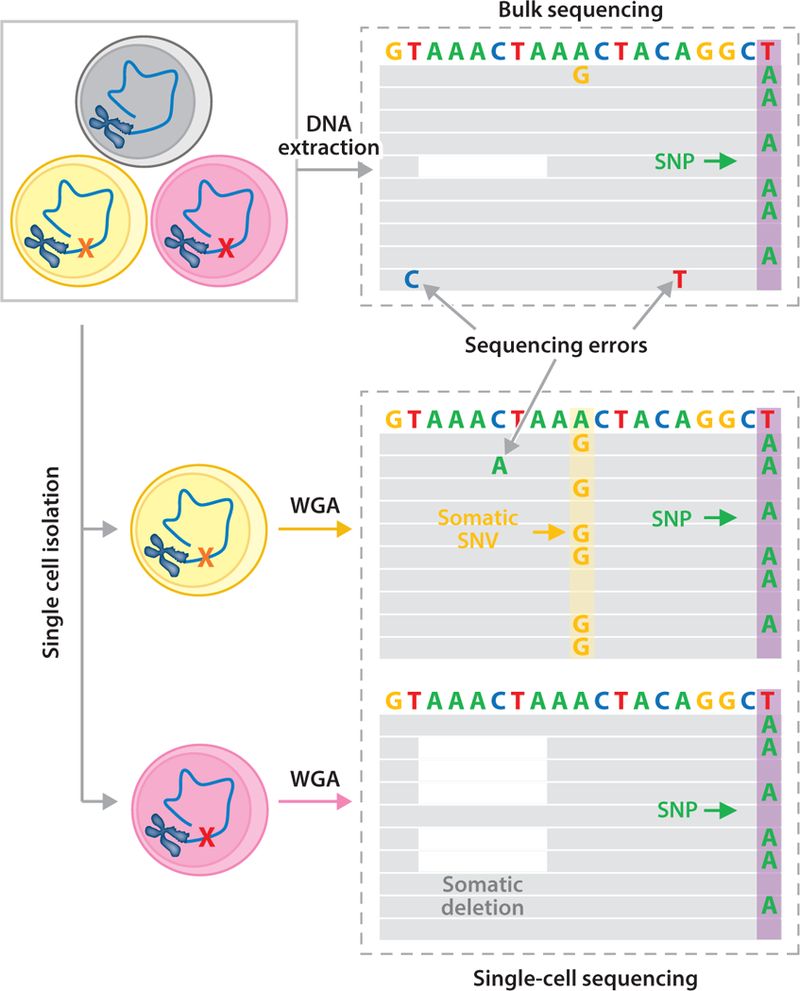
Single-cell analysis detects somatic mutations. Using bulk sequencing, somatic SNVs (e.g., the A→G mutation) and deletions (e.g., the white space) cannot be detected because they are unique to a cell (the yellow and red cells on the left) and, consequently, affect only one out of many thousands of reads. SNVs cannot even be distinguished from sequencing errors, which occur at a much higher frequency than somatic mutations, i.e., 1/100. These low-abundant SNVs and deletions can only be detected using single-cell sequencing, through which a heterozygous mutation will be observed in approximately half of the reads. Both the A→G mutation and the deletion are now easily identified. A germline variant (e.g., the T→A SNP; purple columns) is detectable in all single cells and in the bulk DNA. Abbreviations: SNP, single-nucleotide polymorphism; SNV, single nucleotide variation, WGA, whole-genome amplification.
