Figure 1. Evolution of the human sex chromosomes resulted in four gene classes on the X chromosome.
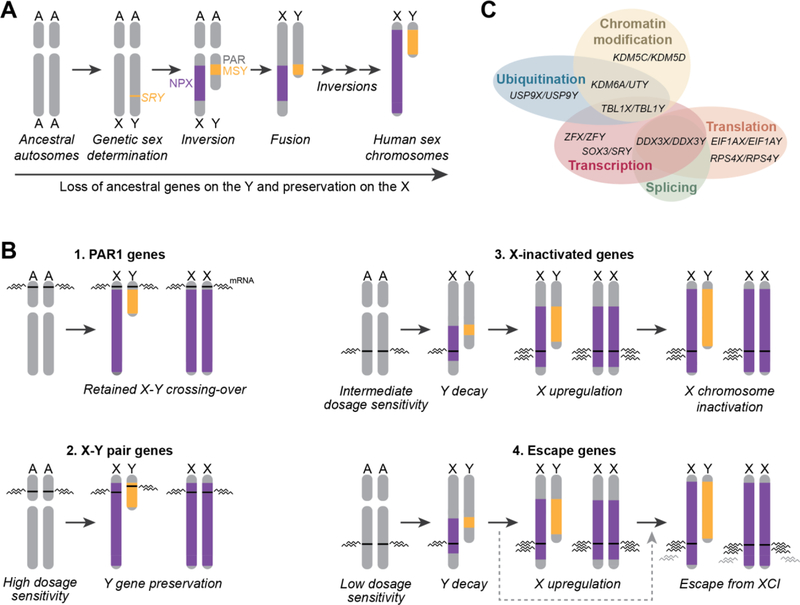
(A) Sex chromosome evolution began with ordinary autosomes. SRY emergence began Y chromosome differentiation, followed by a series of inversions that resulted in loss of crossing-over between the male-specific region of the Y (MSY) and the non-pseudoautosomal region of the X (NPX). The pseudoautosomal region (PAR) on the short arm retained X-Y crossing over. (B) Four classes of genes on the X chromosome underwent different evolutionary trajectories as a result of Y chromosome decay. The black wavy lines indicate the level of mRNA expressed from each gene. PAR1 genes (1) retained X-Y crossing-over and remain expressed from both the X and Y chromosomes. X-Y pair genes (2) reside in the NPX and MSY and do not cross-over, but due to high dosage sensitivity the Y gene was preserved and expression retained on both X chromosomes in 46,XX females. X-inactivated genes (3) had intermediate dosage sensitivity, which allowed for Y chromosome decay. This was followed by upregulation on the X chromosome to retain ancestral dosage in males, and subsequent inactivation of one allele in females. Escape genes (4) followed the same initial path as the X-inactivated genes, but did not become inactivated. Some escape genes may have avoided the step of X-upregulation, indicated by the gray dashed arrow bypassing this step. These genes are predicted to have lower levels of mRNA expression (gray wavy lines) compared to genes that underwent X-upregulation. (C) Functions of select human X-Y pair genes, adapted from (Bellott et al., 2014).
