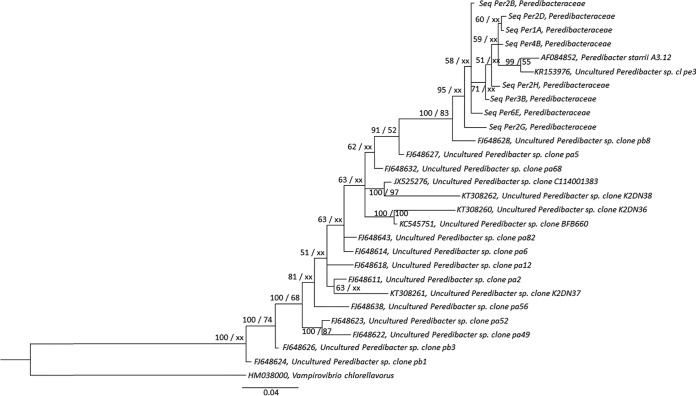
FIG 5.
Bayesian tree generated from 16S rRNA gene data set of Lakes Annecy, Bourget, and Geneva with 8 curated and clustered Peredibacteraceae centroid sequences originating from Sanger sequencing, along with 19 other sequences retrieved from the NCBI (GenBank) (67), including one type species, Peredibacter starrii. These sequences were aligned using MUSCLE (66) via the MEGA6 software (60), and the alignment was curated using Gblocks (68), resulting in 544 positions from 570 positions. A Bayesian tree was built using MrBayes 3.2.6 (72) with 5 million generations and a burn in value of 25% using a GTR+I+G model. As for the ML tree, PhyML-3.1 (71) was used based on a jModelTest-2.1.1 (69) best substitution model, i.e., TrN+I+G. Posterior probability (PP) values followed by bootstrap values are added to the left of a node when possible (PP/BS). Bootstraps below 50 were deleted. Accession numbers are listed to the left of some organism names. Vampirovibrio chlorellavorus was used as an outgroup to root the Peredibacteraceae tree.