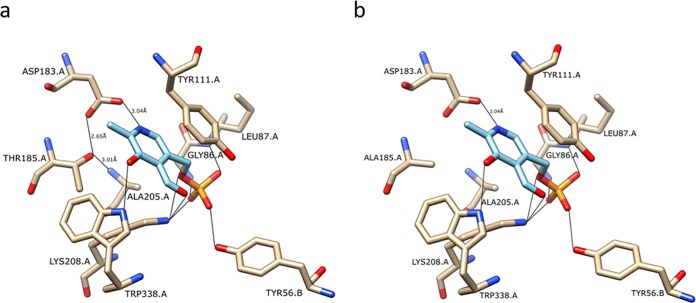
FIG 2.
Model of the Irc7pL active site in the presence of PLP cofactor (light blue). The model shows amino acid residues from two subunits (A and B) of Irc7pL that form the catalytic dimer. In the model on the left (a), the hydroxyl group of Thr185 forms a hydrogen bond with the carboxylate group of Asp183, fixing it for optimal interaction with the nitrogen of the pyridine ring of PLP. In the model on the right (b), mutation on the Thr185 to Ala results in the loss of the hydrogen bond between this amino acid and Asp183.