In this study, the epigenetic global regulator rtfA, which encodes a putative RNA-Pol II transcription elongation factor-like protein, was characterized in the mycotoxigenic and opportunistic pathogen A. flavus. Specifically, its involvement in A. flavus pathogenesis in plant and animal models was studied. Here, we show that rtfA positively regulates A. flavus virulence in both models. Furthermore, rtfA-dependent effects on factors necessary for successful invasion and colonization of host tissue by A. flavus were also assessed. Our study indicates that rtfA plays a role in A. flavus adherence to surfaces, hydrolytic activity, normal cell wall formation, and response to oxidative stress. This study also revealed a profound effect of rtfA on the metabolome of A. flavus, including the production of potent mycotoxins.
Keywords: pathogenicity, rtfA, secondary metabolism, aflatoxin, mycotoxin, genetic regulation, metabolome, Aspergillus flavus
ABSTRACT
Aspergillus flavus is an opportunistic fungal plant and human pathogen and a producer of mycotoxins, including aflatoxin B1 (AFB1). As part of our ongoing studies to elucidate the biological functions of the A. flavus rtfA gene, we examined its role in the pathogenicity of both plant and animal model systems. rtfA encodes a putative RNA polymerase II (Pol II) transcription elongation factor previously characterized in Saccharomyces cerevisiae, Aspergillus nidulans, and Aspergillus fumigatus, where it was shown to regulate several important cellular processes, including morphogenesis and secondary metabolism. In addition, an initial study in A. flavus indicated that rtfA also influences development and production of AFB1; however, its effect on virulence is unknown. The current study reveals that the rtfA gene is indispensable for normal pathogenicity in plants when using peanut seed as an infection model, as well as in animals, as shown in the Galleria mellonella infection model. Interestingly, rtfA positively regulates several processes known to be necessary for successful fungal invasion and colonization of host tissue, such as adhesion to surfaces, protease and lipase activity, cell wall composition and integrity, and tolerance to oxidative stress. In addition, metabolomic analysis revealed that A. flavus rtfA affects the production of several secondary metabolites, including AFB1, aflatrem, leporins, aspirochlorine, ditryptophenaline, and aflavinines, supporting a role of rtfA as a global regulator of secondary metabolism. Heterologous complementation of an A. flavus rtfA deletion strain with rtfA homologs from A. nidulans or S. cerevisiae fully rescued the wild-type phenotype, indicating that these rtfA homologs are functionally conserved among these three species.
IMPORTANCE In this study, the epigenetic global regulator rtfA, which encodes a putative RNA-Pol II transcription elongation factor-like protein, was characterized in the mycotoxigenic and opportunistic pathogen A. flavus. Specifically, its involvement in A. flavus pathogenesis in plant and animal models was studied. Here, we show that rtfA positively regulates A. flavus virulence in both models. Furthermore, rtfA-dependent effects on factors necessary for successful invasion and colonization of host tissue by A. flavus were also assessed. Our study indicates that rtfA plays a role in A. flavus adherence to surfaces, hydrolytic activity, normal cell wall formation, and response to oxidative stress. This study also revealed a profound effect of rtfA on the metabolome of A. flavus, including the production of potent mycotoxins.
INTRODUCTION
The genus Aspergillus is composed of numerous species of medical, industrial, and agricultural importance. Some of these species are opportunistic pathogens, and many produce a variety of secondary metabolites, among them beneficial compounds such as antibiotics, cholesterolemia-reducing drugs, and antitumor compounds. Other Aspergillus secondary metabolites, such as mycotoxins, present detrimental properties. Aspergillus flavus is widely known as an opportunistic fungal pathogen of economically important oil seed crops, contaminating them with mycotoxins, such as the polyketide-derived compounds known as aflatoxins (AFs). Among them, aflatoxin B1 (AFB1) is the most mutagenic and carcinogenic natural compound known (1–4). AFB1 primarily targets the liver, and chronic low-level AFB1 exposure has been shown to cause immunosuppression and hepatocellular carcinoma, among other illnesses (5, 6). Acute AFB1 exposure can lead to aflatoxicosis, which can be lethal (7).
The A. flavus genome has been predicted to contain 56 secondary metabolite gene clusters involved in the production of a wide variety of metabolites (8–10). In addition to AFs, A. flavus can also produce the indole tetramic acid cyclopiazonic acid (CPA) and the indole diterpene aflatrem. CPA is an inhibitor of calcium-dependent ATPase, which leads to altered levels of Ca2+ in the sarcoplasmic reticulum, and aflatrem is widely known to cause neurological disorders due to its tremorgenic properties (11–13). Interestingly, A. flavus is also a producer of beneficial compounds, such as leporins, aflavinines, ditryptophenaline, and aspirochlorine. Leporin B is one of a group of secondary metabolites collectively known as leporins and has been shown to reduce hypoglycemia (14). Aflavinines are sclerotial metabolites with anti-insectan properties (15). In addition, ditryptophenaline is analgesic and anti-inflammatory, and aspirochlorine exhibits antifungal and antibacterial activities (16, 17).
In developed countries, strict legislation has been set to control the maximum amount of total AFs present in food commodities to protect public health (18, 19). In the United States, estimated losses to the corn industry alone due to AF contamination range from $52.1 million to $1.68 billion (19, 20). In developing countries lacking this legislation, human and animal consumption of AF-contaminated crops often leads to illness and, in some cases, death.
In addition to infecting important crops, A. flavus has also been known to cause a deadly lung infection known as invasive aspergillosis (IA). Although A. flavus is the second leading cause of IA, after Aspergillus fumigatus, infections caused by A. flavus are 100-fold more virulent than those caused by A. fumigatus (21–23). A. flavus laboratory animal infections showed fungal biomass accumulating in the liver, lungs, kidneys, and brain (24, 25). The 4- to 6-μm-diameter A. flavus conidia can be deposited in the upper respiratory tract, resulting in upper respiratory infections (23, 26–30). In addition to respiratory infections, A. flavus has also been shown to be a causative agent of other types of human infections, including fungal keratitis, accounting for 80% of cases (23, 31).
Different factors can contribute to the success of A. flavus as an opportunistic pathogen; for example, factors affecting invasion and colonization of the host plant or animal tissue include adhesion to surfaces that is necessary for biofilm formation, extracellular hydrolytic activity, maintenance of cell wall structure, and resistance to oxidative stress. Biofilm production helps the invading microorganism evade host immune responses (32). In addition, A. flavus is known to produce a wide variety of extracellular hydrolytic enzymes, such as proteases, lipases, and amylases. These hydrolytic enzymes contribute to the breakdown of plant or human tissue resulting in colonization of the host (33, 34). Cell wall composition and integrity are relevant to protect fungal cells from environmental insults (35). During invasion and colonization of host tissue, fungi experience a wide variety of different stressors, including oxidative stress, a condition that fungal cells must be able to endure (36).
Due to the devastating impacts of A. flavus pathogenesis on agriculture and health, it is imperative to develop new methodologies to reduce these negative effects, including strategies that target key regulators of fungal virulence. One of these possible targets is the gene known as rtfA. This gene was first characterized in Saccharomyces cerevisiae, where its product was shown to be an RNA polymerase II (Pol II) transcription elongation factor involved in numerous functions, including ubiquitination of histone H2B, dimethylation and trimethylation of histone H3, TATA site selection by TATA box binding proteins (TBP), interactions with active open reading frames (ORFs), proper attachment of components from RNA Pol II, and binding chromatin remodeling proteins, such as the ATP-dependent protein Chd1 (37–43). Homologs of rtfA have been found in Aspergillus species. In Aspergillus nidulans, A. fumigatus, and A. flavus, rtfA regulates the development and production of some secondary metabolites, including AF production in A. flavus (44–46). In A. fumigatus, rtfA is also involved in virulence (45); however, whether the rtfA homolog in A. flavus is relevant in infection of either plants or animals has not been investigated. In this study, we examined the role of the rtfA gene in A. flavus virulence using two model systems, a peanut seed model for plant infection and Galleria mellonella as an IA infection model in animals. Furthermore, we characterized the role of rtfA in cellular processes that are known to be essential for successful invasion, colonization, and survival of A. flavus, specifically, adherence to surfaces, maintenance of cell wall composition, hydrolytic activity, and tolerance to oxidative stress. This study also revealed the profound effect of rtfA on the A. flavus metabolome and assessed possible functional conservation among S. cerevisiae, A. nidulans, and A. flavus rtfA homologs.
RESULTS
RtfA presents low conservation with putative homologs beyond filamentous fungi.
rtfA homologs are present in the genomes of several eukaryotic organisms, including fungi. These homologs contain a conserved Plus3 domain. This domain contains three positively charged amino acids, specifically two arginine residues and a serine residue, that give rise to the name for this domain (47). Studies in humans have shown that the Plus3 domain can bind to single-stranded DNA in addition to RNA polymerase II (47). Previous bioinformatic analyses revealed that in Aspergillus spp., as well as in other Ascomycetes, putative rtfA homologs could also be present (44). In this study, BLASTP analysis indicated a high conservation of the A. flavus RtfA protein with those of A. nidulans (70.4% identity and 82.1% similarity) and A. fumigatus (75.2% identity and 85.6% similarity). However, the conservation with S. cerevisiae Rtf1 is low (25% identity and 44% similarity). Previously, Warner et al. (43) compared the S. cerevisiae Rtf1 amino acid sequence to homologs in Schizosaccharomyces pombe, Homo sapiens, and Caenorhabditis elegans and also found low levels of conservation between these sequences. To examine the conservation of A. flavus RtfA with those of higher eukaryotic species, we performed a BLASTP and box shade analysis. Our results revealed that the A. flavus RtfA amino acid sequence also showed low conservation (19.5% to 21.5% identity and 33.9% to 37.9% similarity) with putative homologs in higher eukaryotic species (see Fig. S1 in the supplemental material).
Heterologous complementation of ΔrtfA mutant with homologs in model fungi, A. nidulans and S. cerevisiae, rescues wild-type phenotype.
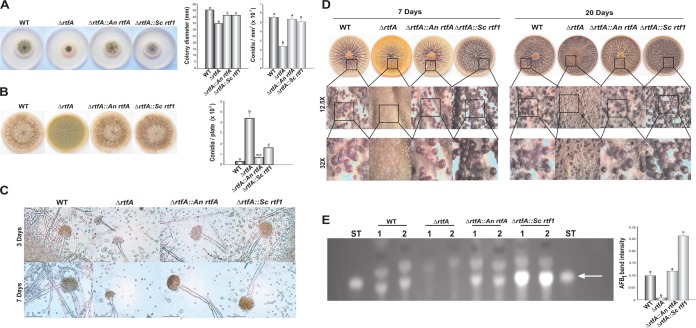
To determine whether putative homologs of rtfA from S. cerevisiae and A. nidulans are functionally conserved, we generated heterologous complementation strains using rtf1 and rtfA corresponding wild-type alleles and transformed them in the A. flavus rtfA deletion strain. Transformants were confirmed using diagnostic PCR (Fig. S2A and B). The heterologously complemented strains carrying the A. nidulans rtfA and S. cerevisiae rtf1 wild-type alleles were assessed with respect to their vegetative growth, asexual development, sclerotial and AFB1 production, and cellular processes previously described to be influenced by rtfA in A. flavus (46). The colony diameters of the heterologously complemented strains were larger than that of the ΔrtfA mutant strain, indicating fungal growth recovery by the homologous genes (Fig. 1A). In addition, the heterologous complementation strains did not present statistically significantly different conidiation levels from those of the wild-type strain at the early time point, but these were statistically significantly different from those of the ΔrtfA mutant strain (Fig. 1A). However, at a later time point, conidial production in the strain with the A. nidulans rtfA heterologous complementation was similar to that in the wild type and statistically significantly lower than that of the A. flavus ΔrtfA mutant strain (Fig. 1B). Conidiophore vesicle development, previously described to be affected by rtfA (46), was also evaluated. Both heterologous complementation strains exhibited conidiophore vesicle diameters similar in size to the wild type and larger than those in the ΔrtfA mutant strain (Fig. 1C). With respect to sclerotial development, the absence of rtfA repressed sclerotial formation (Fig. 1D) (46). Complementation with A. nidulans rtfA or S. cerevisiae rtf1 restored wild-type sclerotial production at both time points assayed (Fig. 1D).
FIG 1.
The function of rtfA homologs of A. flavus, A. nidulans, and S. cerevisiae is conserved. (A) Photographs of point-inoculated cultures of wild-type (WT) and ΔrtfA, ΔrtfA::An rtfA, and ΔrtfA::Sc rtf1 strains after 3 days of incubation at 30°C in the dark. On the right, measurement of vegetative growth as colony diameter and conidial quantification. (B) Images and conidial quantifications of the A. flavus cultures after 11 days of incubation at 30°C in the dark. (C) Micrographs of the A. flavus conidiophores after 3 days and 7 days of incubation at 30°C in the dark. (D) Analysis of sclerotial production in the A. flavus strains after 7 and 20 days of incubation on Wickerham medium at 30°C in the dark. (E) TLC analysis of AFB1 levels and corresponding densitometry after 7 days of incubation at 30°C in the dark. Arrow indicates AFB1 in TLC image. Error bars represent the standard error. Columns with different letters represent values that are statistically different (P < 0.050).
AFB1 production was also analyzed in these strains. The strain complemented with A. nidulans rtfA showed toxin levels similar to those in the wild type (Fig. 1E). The S. cerevisiae rtf1 heterologous complementation strain rescued AFB1 synthesis, producing statistically significantly higher levels of AFB1 than the control.
rtfA is indispensable for A. flavus pathogenesis on live plant and animal tissue.
Previously, rtfA was extensively characterized in the opportunistic pathogen A. fumigatus, where it was found to affect virulence (45). Based on this and on the high conservation observed in the RtfA deduced amino sequence in both fungi, we hypothesized that rtfA could also regulate virulence in A. flavus. For this reason, the pathogenicity of A. flavus in both plant seeds and animals was assessed in the presence and absence of rtfA.
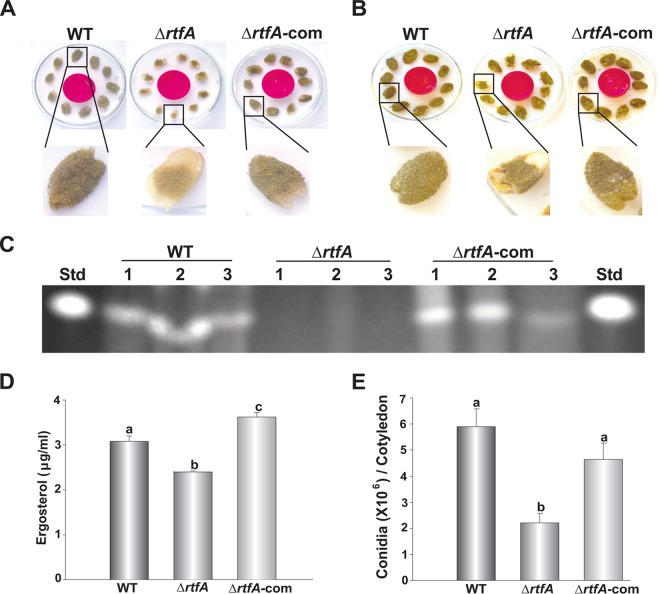
To determine whether rtfA plays a role in plant pathogenesis in A. flavus, viable peanut cotyledons were infected with the wild-type, ΔrtfA mutant, and rtfA complementation (ΔrtfA-com) strains. Cultures were photographed at 3 and 5 days of incubation (Fig. 2A and B). Our study revealed a complete abolishment of AFB1 production in viable seeds infected with the ΔrtfA mutant strain (Fig. 2C). In this experiment, levels of a fungus-specific sterol known as ergosterol were used as an indicator of fungal burden present in the infected plant tissue (Fig. 2D). Seeds infected with the ΔrtfA mutant strain contained significantly less ergosterol than did seeds infected with the control strains. In addition, the absence of rtfA resulted in a statistically significant decrease in conidial production (Fig. 2A, B, and E).
FIG 2.
rtfA is required for normal pathogenicity during Aspergillus flavus infection of peanut seeds. Aspergillus flavus wild-type (WT), rtfA deletion (ΔrtfA) mutant, and rtfA complementation (ΔrtfA-com) strains were inoculated on the sterilized adaxial cotyledon surfaces of NC94022 peanut seeds and incubated for 72 h (A) and 120 h (B). (C) TLC analysis of aflatoxin B1 content in the infected seeds after 5 days of incubation. (D) HPLC quantification of fungal ergosterol content in the infected seeds. (E) Quantification of conidial production. Error bars represent the standard error. Columns with different letters represent values that are statistically different (P < 0.050).
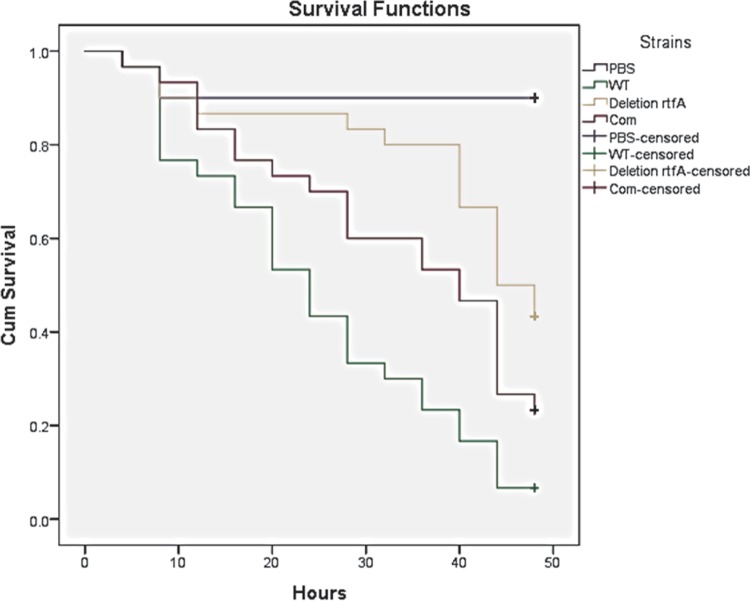
Since A. flavus is also an opportunistic human and animal pathogen, we evaluated the role of rtfA in a well-known animal infection model organism used for invasive aspergillosis studies, G. mellonella. Thirty G. mellonella larvae were infected per fungal strain, and survival rates were monitored for 48 h after an initial 24-h postinfection period. The results of this analysis revealed a statistically significant decrease in mortality rates in the larvae infected with the ΔrtfA mutant strain in comparison to those infected with the wild-type and ΔrtfA-com strains (Fig. 3).
FIG 3.
rtfA is necessary for normal virulence in the animal model Galleria mellonella. The A. flavus strains were used to infect G. mellonella larvae. Thirty replicates were used per strain. Thirty animals were also used as controls, injected with PBS buffer only. An additional control with 10 noninjected animals was used. The animals were maintained at 30°C in the dark for 24 h. After that, survival rates were monitored every 4 h. Data representation was carried out using SPSS statistical software. Cum, cumulative.
Absence of rtfA affects the ability of A. flavus to adhere to surfaces.
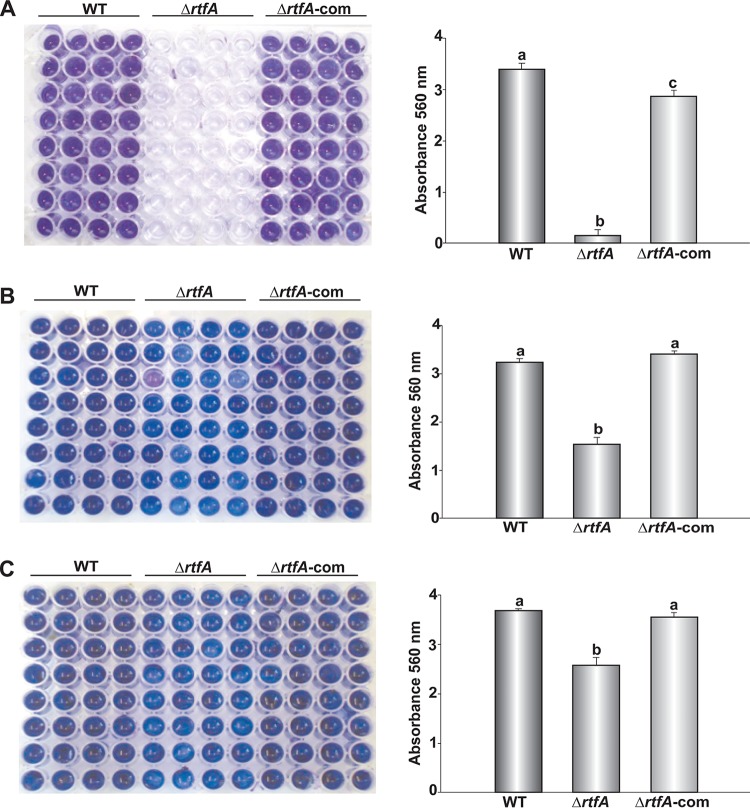
Biofilm formation is a characteristic shared by many pathogenic microorganisms, including fungi. Species of fungi from the genus Aspergillus, including A. flavus, have been shown to be capable of producing biofilms (48). In addition, 65% of fungal infections in humans are biofilm associated (48). One factor necessary for successful biofilm formation is the ability of the microorganism to adhere to surfaces (49). Previously, A. fumigatus rtfA was shown to influence the ability of this fungus to adhere to surfaces in the absence of rtfA, causing a delay in this process (45). In the current study, adhesion of wild-type, ΔrtfA mutant, and ΔrtfA-com strains was tested at 24 h, 48 h, and 72 h. The results showed a statistically significant decrease and delay in the ability of the A. flavus ΔrtfA mutant strain to adhere to surfaces with respect to the controls (Fig. 4).
FIG 4.
rtfA is necessary for A. flavus normal adhesion to surfaces. Adherence was assessed as mentioned in Materials and Methods, by obtaining absorbance readings after at 560 nm after 2 days (A), 3 days (B), and 4 days (C). Darker blue coloring corresponds to greater adherence. Error bars represent the standard error. Columns with different letters represent values that are statistically different (P < 0.050).
Enzymatic activity in A. flavus is positively regulated by rtfA.
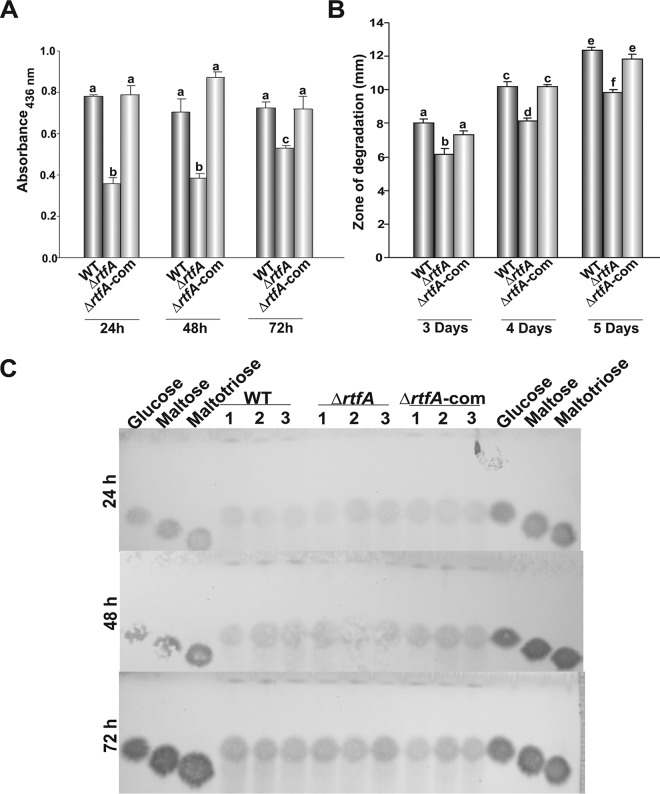
Aspergillus flavus uses several cellular processes to successfully invade and colonize the host, including hydrolytic activity (33, 34). Here, we determined whether rtfA affects protease, lipase, and amylase activities in A. flavus. A statistically significant decrease in protease production was observed in the absence of rtfA after 24 h, 48 h, and 72 h of incubation compared to the control (Fig. 5A). Lipase activity was also reduced in the A. flavus ΔrtfA mutant strain compared to the isogenic control strains (Fig. 5B). In the case of amylase activity, no differences were observed among the strains tested (Fig. 5C).
FIG 5.
Protease and lipase activities in A. flavus are rtfA dependent. (A) Protease activity from GMM plus BSA liquid shaking cultures after 24, 48, and 72 h of incubation, measured using an azocasein-based assay. (B) Lipase activity assessed in A. flavus cultures using tributyrin medium. Zones of degradation were measured after 3, 4, and 5 days of incubation. (C) Analysis of amylase activity induced in PBS medium containing starch. Enzyme activity was assessed using maltoheptose as the substrate and observing degradation products by TLC analysis after 24, 48, and 72 h of incubation. Error bars represent the standard error. (A and B) Columns with different letters represent values that are statistically different (P < 0.050).
rtfA is necessary for normal cell wall composition and integrity in A. flavus.
The cell wall of fungi is an important component of fungal cells that provides structural integrity. Most importantly, the fungal cell wall acts as a barrier to hostile environments, a storage container for enzymes and dangerous compounds, and a structure that is useful in the penetration of live or dead substrates (35). SDS is a common compound used to test fungal cell wall integrity (50, 51). Interestingly, when the A. flavus strains were grown on yeast-glucose-trace elements (YGT) medium containing 0.01% SDS, a statistically significant increase in the sensitivity to this compound was observed in the ΔrtfA mutant compared to that in the isogenic control strains, as measured by percent reduction of growth compared to the growth of each strain on YGT medium not supplemented with SDS (Fig. 6).
FIG 6.
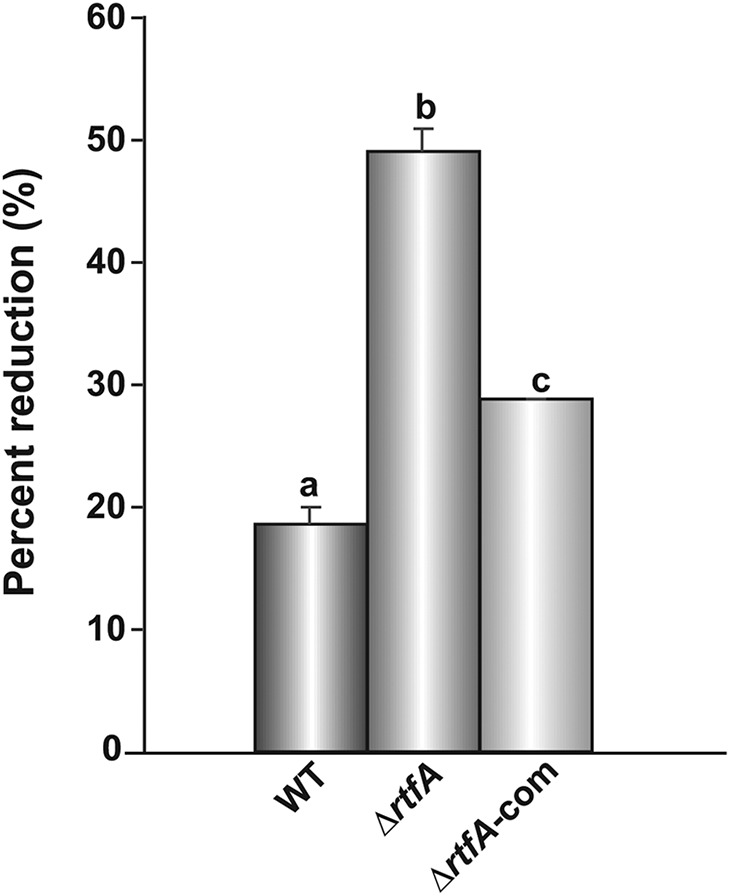
rtfA influences cell wall integrity in A. flavus. The A. flavus strains were point-inoculated on YGT medium and YGT medium containing 0.01% SDS. Cultures were incubated at 30°C in the dark for 72 h. Colony diameter was measured after incubation. The experiment was carried out in duplicate. Data are represented as the percentage of reduction in the growth of strains on YGT medium versus YGT medium containing 0.01% SDS. Error bars represent the standard error. Columns with different letters represent values that are statistically different (P < 0.050).
It is possible that this sensitivity to SDS could be due to defects in cell wall composition in the absence of rtfA. To test this hypothesis, mannoprotein, chitin, and glucan content in soluble and insoluble alkali was measured in the wild-type, ΔrtfA mutant, and ΔrtfA-com strains. Our results revealed no differences in mannoprotein levels. However, a statistically significant decrease in chitin levels was observed. In addition, an increase of glucan in the alkali-soluble fraction accompanied by a decrease of the same in the alkali-insoluble fraction was detected in the ΔrtfA mutant strain in comparison to levels in the control strains (Table 1).
TABLE 1.
Cell wall composition
| Strain | Amt (mean ± SE) (μg/mg) ina
: |
|||
|---|---|---|---|---|
| Alkali-soluble fraction |
Alkali-insoluble fraction |
|||
| Mannoprotein | Glucan | Glucan | Chitin | |
| WT | 5.18 ± 0.07 A | 2.03 ± 0.08 A | 2.45 ± 0.20 A | 4.44 ± 0.41 A |
| ΔrtfA mutant | 5.05 ± 0.15 A | 2.74 ± 0.07 B | 2.05 ± 0.07 A | 3.05 ± 0.26 B |
| ΔrtfA-com strain | 5.02 ± 0.09 A | 2.10 ± 0.06 A | 2.52 ± 0.12 A | 3.64 ± 0.25 AB |
Different uppercase letters represent a significant difference.
rtfA is indispensable for oxidative stress tolerance.
Fungi are robust organisms that can survive exposure to a wide variety of biotic and abiotic stresses. In this study, we specifically examined whether rtfA is necessary for resistance to oxidative stress using menadione. The A. flavus strains were exposed to various concentrations of this compound. After 72 h of incubation, no growth was observed in the rtfA deletion strain in the presence of 0.4 mM menadione, while the control strains were able to form colonies (Fig. 7).
FIG 7.
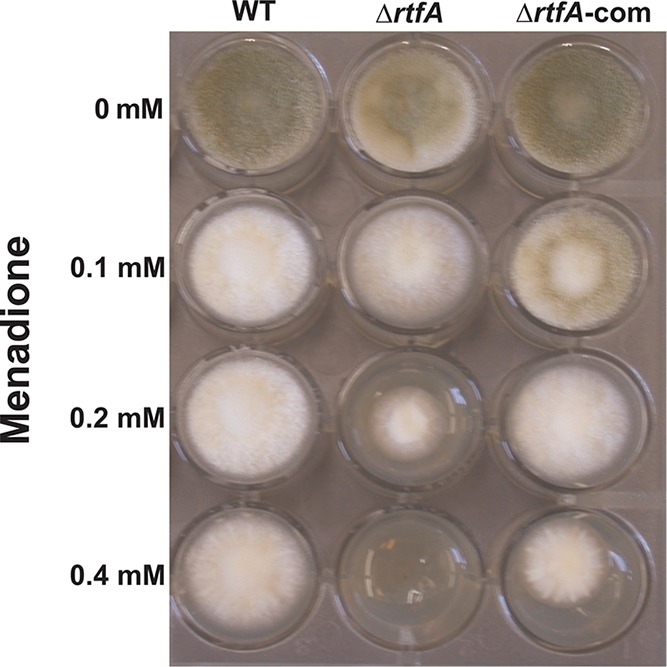
rtfA affects oxidative stress resistance in A. flavus. Two milliliters of YGT agar medium containing different concentrations of menadione was placed into a 24-well plate. Strains were point-inoculated and allowed to grow for 72 h at 30°C in the dark.
rtfA regulates the production of numerous secondary metabolites in A. flavus.
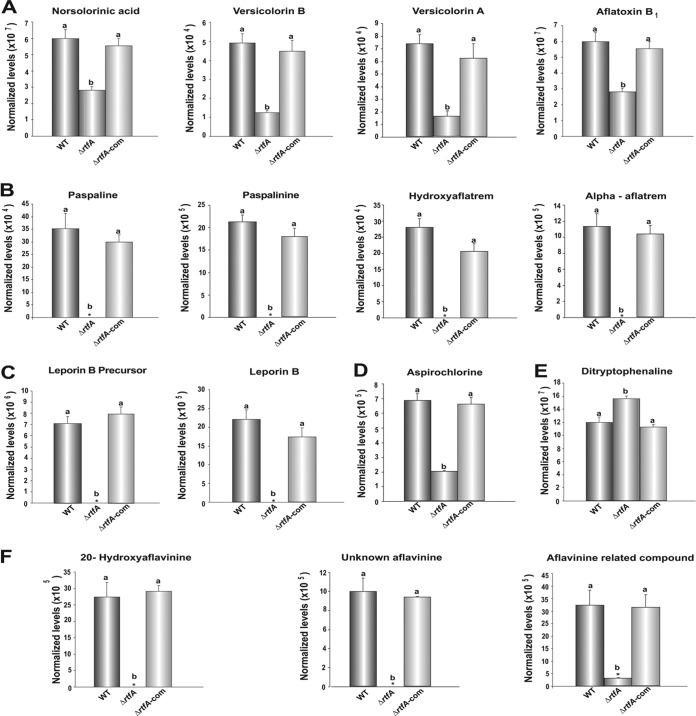
Aspergillus flavus has the capability to produce a wide variety of bioactive compounds with detrimental or beneficial effects on health. Using liquid chromatography-mass spectrometry (LC-MS), we elucidated the rtfA-dependent metabolome in A. flavus. Specifically, our analysis revealed that the production of norsolorinic acid, versicolorin B, and versicolorin A (intermediates in the AF biosynthetic pathway) and the final product AFB1 was positively regulated by rtfA (Fig. 8A). Only O-methylsterigmatocystin was unaffected by absence of rtfA (Fig. S3A). In addition, the rtfA mutant was unable to produce paspaline, paspalinine, hydroxyaflatrem, and alpha-aflatrem, from the aflatrem biosynthesis pathway, under conditions that allowed the production of these compounds in the controls (Fig. 8B). In addition, a leporin B precursor and leporin B were absent in the ΔrtfA mutant strain, while they were detected in the controls (Fig. 8C). Interestingly, decreased levels of aspirochlorine and significantly elevated levels of ditryptophenaline were observed in the ΔrtfA mutant strain compared to those in the wild-type and ΔrtfA-com strains (Fig. 8D and E). Furthermore, a number of compounds believed to be intermediates or end products of the aflavinine biosynthetic pathway, including 20-hydroxyaflavinine, an unknown aflavinine compound (C28H39NO2, m/z 404.29 for [M+H]+), and an aflavinine-related compound (C28H37NO4, m/z 434.27 for [M-H2O+H]+ and m/z 452.28 for [M+H]+ and 474.26 for [M+Na]+) previously described as compound 10 in a study by Forseth et al. (52), were absent in the ΔrtfA mutant strain extracts (Fig. 8F). Our study also revealed that the production of compounds by the cyclopiazonic acid (CPA) pathway, specifically, 2-oxo-CPA and the final product CPA, was unaffected in the absence of rtfA (Fig. S3B).
FIG 8.
rtfA regulates secondary metabolite production in A. flavus. A. flavus wild-type (WT), ΔrtfA mutant, and ΔrtfA-com strains were grown in YGT medium for 5 days at 30°C in the dark. Compounds in the culture supernatants were analyzed by LC-MS. Relative quantification of metabolites in the biosynthetic pathway of aflatoxin B1 (A) and of aflatrem (B). Analysis of metabolites in the biosynthesis pathway of leporins (C), aspirochlorine (D), ditryptophenaline (E), and aflavinines (F). Error bars represent standard error. Columns with different letters represent values that are statistically different (P < 0.050).
DISCUSSION
Aspergillus flavus is an agriculturally and medically relevant opportunistic pathogen. For this reason, it is imperative to identify potential genetic targets that can be used in novel strategies to reduce A. flavus survival, dissemination, production of toxic compounds, and virulence. Among these novel genes is rtfA, a putative RNA Pol II transcription elongation factor. Putative homologs of RtfA containing a Plus3 domain have been found in nonfungal eukaryotes (47, 53, 54); however, based on our study, the conservation between those homologs and A. flavus RtfA is limited. These differences could reflect a significant evolutionary rewiring of this epigenetic factor. The conservation is also moderate compared to Rtf1 in the yeast model S. cerevisiae (44); however, it is highly conserved among other Aspergillus spp., including A. nidulans and A. fumigatus. Some distinct rtfA-dependent effects were observed in different Aspergillus spp., however. For example, reduction of vegetative growth was observed in the A. nidulans, A. fumigatus, and A. flavus ΔrtfA mutant; however, this decrease was minor in A. flavus (44–46). Also, the absence of rtfA affected conidiation in the three Aspergillus spp., but differently. In A. nidulans, the deletion of rtfA resulted in a reduction of conidiation, while in A. fumigatus, it causes hyperconidiation (44, 45), and in A. flavus, a loss of rtfA function resulted in an initial delay in the onset of conidiation, followed by hyperconidiation over time (46). The rtfA homologs also present commonalities in the regulation of cleistothecial and sclerotial formation in A. nidulans and A. flavus, respectively; in both cases, a lack of rtfA resulted in impaired production of these structures (44, 46). In addition, rtfA has been shown to be a positive regulator of the production of several secondary metabolites in A. nidulans, including sterigmatocystin (44), and in A. flavus, rtfA is necessary for the synthesis of AFB1 (46), suggesting a common regulatory mechanism, whereas in A. fumigatus, rtfA negatively controls secondary metabolism (45).
The RtfA homolog Rtf1 in S. cerevisiae was previously shown to be a component of the Paf1 complex (37–43). In spite of the difference between the amino acid sequences of Rtf1 and A. fumigatus RtfA, complementation of the A. fumigatus ΔrtfA mutant strain with rtf1 recovered the wild-type phenotype, suggesting similar mechanisms for the two homologs. As mentioned above, RtfA homologs present a Plus3 domain shown to bind to RNA polymerase II as well as to single-stranded DNA from yeast to humans (47). To elucidate possible functional conservation of A. flavus RtfA with those from other organisms, we heterologously complemented the A. flavus ΔrtfA mutant with putative homologs from two model fungi, A. nidulans and S. cerevisiae. Both heterologous complementations rescued many traits of the wild-type phenotype in terms of growth, conidiation, sclerotial production, and ABF1 production, suggesting that these homologs are functionally conserved.
Mutations in rtf1 or rtfA result in pleiotropic effects in S. cerevisiae, A. nidulans, A. fumigatus, and A. flavus (37–46). In yeast, Rtf1 is an epigenetic regulator involved in chromatin remodeling by several mechanisms. For example, Rtf1 controls monoubiquitination of histone H2B at lysine 123, required for the methylation of histones H3K4 and H3K79 (37, 40, 42, 43). Rtf1 has been shown to regulate gene silencing of subtelomeric regions through methylations of histone H3 by the Set1-containing COMPASS and Dot1 at lysines K4 and K79, respectively (40, 55, 56). Due to the fact that histone modifications modulate the expression of numerous genes (57–59), it is expected that the absence of Rtf1/RtfA could result in the observed pleiotropic effects, including changes in virulence.
Importantly, the current study demonstrated that rtfA is relevant in A. flavus pathogenesis using a plant model and an animal model. Peanut seed infected with the deletion strain showed a reduction in conidial production compared to seed infected with the wild-type strain. Additionally, the ΔrtfA mutant failed to produce AFB1 and demonstrated reduced seed colonization, as determined by ergosterol levels compared to the control. A reduction in virulence in the absence of rtfA was also observed when the G. mellonella animal infection model was used, indicating that rtfA is indispensable for A. flavus virulence. Similarly, rtfA is also relevant in A. fumigatus animal infections (45). It is possible that rtfA homologs could also play an important role during infection in other pathogenic fungi.
In order to gain further insight into the role of A. flavus rtfA in pathogenicity, we assessed several factors postulated to be involved in successful invasion and colonization of host tissue. It was found that the absence of rtfA causes a delay and reduction in the ability of the fungus to adhere to surfaces. This is in agreement with previous observations in A. fumigatus (45), although the delay was not as pronounced as in A. flavus. Attachment of the fungus to surfaces at an early stage of the infection process is a necessary step for the production of biofilm and subsequent evasion of the host immune/defense system response (49). It is likely that the delay and reduction in adherence in the ΔrtfA mutant strains could contribute to the observed decrease in virulence in both A. flavus and A. fumigatus rtfA mutants.
In addition, our results revealed that rtfA positively regulates hydrolytic activity in A. flavus. Specifically, the absence of rtfA resulted in a decrease in protease and lipase activity. Extracellular enzymes, such as proteases and lipases, are relevant to degrade host tissue, allowing the fungus to thrive inside the host (33, 34). It is possible this decrease in enzymatic activity could prevent the rtfA mutant from successfully colonizing the host, as suggested by the reduction in ergosterol levels of seed infected with the mutant compared to the wild type. The reduction in lipase activity is particularly relevant in the case of A. flavus colonization of oil-rich seed crops.
Previously, rtfA was shown to be required for normal fungal growth when A. flavus was exposed to light and high temperature, as at 42°C under light conditions the rtfA mutant presented a reduction in colony growth compared to the wild type (46). Higher sensitivity to elevated temperatures could be caused by defects in the cell wall (60); in this case, possible rtfA mutant-associated defects in the cell wall could then result in higher temperature sensitivity leading to growth reduction. The present study demonstrates that the absence of rtfA also results in greater sensitivity to SDS. This further supports the possibility of rtfA-dependent defects in the cell wall. Indeed, although mannoprotein levels were not affected by rtfA, a statistically significant decrease in chitin was observed in the strain lacking rtfA compared to the wild type, indicating that rtfA positively influences the synthesis of this polymer in A. flavus. It has been shown that chitin has specific immunological effects, for example, the activation of certain types of immune cells, such as peritoneal/alveolar macrophages and natural killer cells in mammals (61), that could be decreased in the absence of rtfA. The ratio of glucan present in the alkali-soluble and -insoluble fractions of the cell wall was altered in the A. flavus rtfA mutant. Like chitin, fungal β-glucan levels of a host can lead to the activation of an immune response (62). The decrease in chitin together with changes in the glucan ratio could also have weakened the integrity of the cell wall leading to the observed reduction in virulence in the A. flavus rtfA mutant.
Host immune cells such as macrophages and neutrophils produce reactive oxygen species (ROS), which in turn can have deleterious effects on fungal cells (63). Our findings revealed elevated sensitivity of the A. flavus rtfA mutant to oxidative stress in the presence of menadione with respect to the control. This result is comparable to the effect of ROS on an A. fumigatus rtfA mutant (45), indicating that rtfA plays a role in resistance to oxidative stress in these two Aspergillus species.
The fact that the rtfA gene influences multiple cellular processes, including virulence, suggests that this gene and its gene product could be used as a potential novel target to reduce the detrimental effects of A. flavus. For example, RNA interference (RNAi) technology could be utilized to silence rtfA. This technique has recently been used to successfully silence other A. flavus genes expressing RNAi constructs in maize (see, e.g., reference 64). Additionally, small peptides could also be designed to interfere with RtfA-protein interactions, for instance, those between RtfA and other members of the Paf1 complex (39), which could decrease virulence in plants and animals as well as reduce toxin production.
Substrates colonized by A. flavus can become contaminated with secondary metabolites such as AFB1 and CPA. Previously, we reported that rtfA regulates the synthesis of AFB1 in this fungus (46), and the production of other uncharacterized metabolites also appeared to be affected. In other Aspergillus spp., rtfA also affected the biosynthesis of secondary metabolites (44, 45). Specifically, in A. nidulans, the production of sterigmatocystin and penicillin biosynthesis was positively regulated by rtfA (44), while the production of tryptoquivaline F, pseurotin A, fumiquinazoline C, festuclavine, and fumigaclavines A, B, and C was negatively regulated by rtfA in A. fumigatus. In the current study, we analyzed the rtfA-dependent metabolome and found that not only the synthesis of AFB1, but also that of aflatrem, leporins, aflavinines, ditryptophenaline, and aspirochlorine, is controlled by rtfA. Collectively, the results from this and previous studies indicate that rtfA is a global regulator of secondary metabolite production in Aspergillus species.
In conclusion, this study provides insight into the role of rtfA in A. flavus pathogenesis and factors necessary for normal fungal invasion and colonization of the host, including adhesion, hydrolytic activity, cell wall composition, and oxidative stress resistance. Furthermore, we showed the broad regulatory effects of rtfA on the metabolome of A. flavus, including its involvement in the production of potent mycotoxins. This together with the strong effect of rtfA on A. flavus morphogenesis makes rtfA and its gene product RtfA potential targets of strategies to reduce virulence and control mycotoxin contamination in food and feed crops. The fact that rtfA homologs used in this study displayed functional conservation with A. flavus rtfA suggests that such strategies could be implemented against other fungal pathogens.
MATERIALS AND METHODS
Bioinformatic analysis.
Gene and corresponding deduced amino acid sequences for A. flavus RtfA (NCBI RefSeq accession no. XP_002377748.1) were obtained from the NCBI (https://www.ncbi.nlm.nih.gov/). The BLASTP search tool was used to identify homologs in other eukaryotic species. MAFFT sequence alignment (http://www.ebi.ac.uk/Tools/msa/mafft/) was used with all sequences. This was followed by shading of the alignment using the BoxShade server (http://www.ch.embnet.org/software/BOX_form.html).
Strains and culture conditions.
The A. flavus L morphotype strains utilized in this study, unless specified differently, were the CA14 pyrG-1 (pyrG+ niaD− Δku70) control strain, rtfA deletion strain (ΔrtfA, tJML1.1), and rtfA complementation strain (ΔrtfA-com, tJML2.1) (46) (Table 2). All strains were grown on YGT medium (per liter, 20 g glucose, 5 g yeast extract, 1 ml of trace elements [65] with agar [15 g/liter]), unless specified differently. Fungal strains were maintained in 30% glycerol stocks at −80°C.
TABLE 2.
Strains used in this study
| A. flavus strain name | Pertinent genotypea | Reference or source |
|---|---|---|
| CA14 | pyrG− niaD− Δku70 | SRRC collection #1709 |
| CA14 pyrG-1 | pyrG+ niaD− Δku70 | 79 |
| tJML1.1 | ΔrtfA pyrG+ niaD− Δku70 | 46 |
| tJML2.1 | ΔrtfA pyrG+ niaD− Δku70 rtfAA. fla+ | 46 |
| tJML4.1 | ΔrtfA pyrG+ niaD− Δku70 rtfAA. nid+ | This study |
| tJML5.2 | ΔrtfA pyrG+ niaD− ptrA+ Δku70 rtfAS. cer+ | This study |
A. fla, A. flavus; A. nid, A. nidulans; S. cer, S. cerevisiae.
Heterologous complementation of rtfA.
(i) Complementation of the A. flavus ΔrtfA mutant strain with the A. nidulansrtfA gene. In order to generate the heterologous complementation strain with A. nidulans rtfA, a 3.545-kb A. nidulans rtfA fragment was PCR amplified with primers 744 and 1902 (Table 3) using pSM3-rtfAcom (44) as the template. The PCR product was then digested with KpnI and ligated to the pPTR1 vector (TaKaRa, Mountain View, CA, USA) previously digested with the same enzyme. pPTRI contains the Aspergillus oryzae pyrithiamine resistance gene (ptrA) as a selection marker for fungal transformation. The resulting recombinant vector, pJML1.1, was transformed in the A. flavus ΔrtfA mutant strain. Transformants were confirmed by diagnostic PCR with primers 744 and 1902 (Table 3). The selected transformant was designated tJML4.1.
TABLE 3.
Primers used in this study
| Primer name | Sequence (5′ – 3′) |
|---|---|
| 744: rtfA-Com2 | AAAAAATGGTACCTTAGGCAGTGGGTATGATGTTGG |
| 1902: rtfA A. nid kpnI F | AAAAAAGGTACCTCGCAAGCATATCCTTCAACT |
| 1949: A. fla rtfA 5′UTR F P1 | CATCCCGAATGGTACCCCTTGC |
| 1950: A. fla rtfA 5′UTR R P2 | AGGTGGCTGAGTCAGTCGGAAAG |
| 1951: A. fla rtfA 3′UTR F P3 | CGCATTGCTACATGCCGAAGTTCAC |
| 1952: A. fla rtfA 3′UTR R P4 | AAAAAGGTACCTGGGCCTGATGAACCAGTTGCATA |
| 1953: S. cer rtf1 A. flav link F P5 | CTTTCCGACTGACTCAGCCACCTATGTCTGATTTAGATGAGGATTTATTAGCCTTG |
| 1954: S. cer rtf1 A. flav link R P6 | GTGAACTTCGGCATGTAGCAATGCGCTAAAACTTAAGGTCAAATTGATATCCAATTCACC |
| 1955: S. cer A. fla rtfA nest P7 | AAAAAGGTACCAGAGCCAATCTCCGTCTCCACAG |
| 1956: S. cer A. fla rtfA nest P8 | AAAAAGGTACCCGTTTCTGAGCAACAAGGGCAAGG |
(ii) Complementation of the A. flavus ΔrtfA mutant strain with the S. cerevisiaertf1 gene. In order to generate the heterologous complementation strain with S. cerevisiae rtf1, a fusion PCR product containing the coding sequence of rtf1 from S. cerevisiae and the 5′ untranslated region (5′UTR) and 3′UTR of A. flavus rtfA was generated as described by Szewczyk et al. (66). Briefly, the 3′UTR and 5′UTR of A. flavus rtfA were PCR amplified from CA14 genomic DNA using primers 1949 and 1950 and primers 1951 and 1952 (Table 3), respectively. The resulting 1.368-kb and 1.537-kb PCR products were fused to a 1.677-kb fragment corresponding to the S. cerevisiae rtf1, previously PCR amplified from genomic DNA of the S. cerevisiae Y2HGold strain (Clontech, Mountain View, CA, USA) using primers 1953 and 1954. Primers 1955 and 1956 were used to fuse the three fragments, resulting in a 4.049-kb PCR product. This fusion cassette was then digested with KpnI and ligated to pPTR1 previously digested with the same enzyme, generating pJML1.2. This plasmid was then transformed in the A. flavus ΔrtfA mutant strain. Transformants were confirmed with diagnostic PCR using primers 1953 and 1954 (Table 3). The selected transformant was designated tJML5.2.
(iii) Assessment of vegetative colony growth. The A. flavus wild type and ΔrtfA mutant strains, the A. nidulans rtfA heterologous complementation strain (ΔrtfA::An rtfA), and the S. cerevisiae rtf1 heterologous complementation strain (ΔrtfA::Sc rtf1) were point-inoculated on YGT medium and incubated at 30°C in the dark for 3 days. Images of cultures were captured with a Cybershot DSC-W120 camera (Sony, New York, NY, USA). Vegetative colony growth was evaluated as colony diameter (in millimeters). This experiment was carried out in triplicate.
(iv) Asexual development analysis. The A. flavus wild-type and ΔrtfA, ΔrtfA::An rtfA, and ΔrtfA::Sc rtf1 strains were point-inoculated on YGT medium. Conidiation was assessed at 3 and 11 days of incubation. Cores of approximately 7 mm were taken 1 cm away from the center of the colony. Samples were homogenized in water, and conidia were quantified with a hemocytometer (Hausser Scientific, Horsham, PA, USA) and an Eclipse E-400 bright-field microscope (Nikon, Inc., Melville, NY, USA).
An additional experiment was also carried out to observe conidiophore vesicles. Briefly, the A. flavus strains were top-agar inoculated on YGT medium and allowed to incubate at 30°C under dark conditions for 3 days. Micrographs were acquired using an E-600 bright-field microscope (Nikon, Inc., Melville, NY, USA) attached to a Nikon DXM 1200 digital camera.
(v) Sclerotial production assay. In order to determine if heterologous complementation of rtfA restored wild-type sclerotial levels, the A. flavus wild-type and ΔrtfA, ΔrtfA::An rtfA, and ΔrtfA::Sc rtf1 strains were point-inoculated on Wickerham agar medium (per liter, 2 g yeast extract, 3 g peptone, 5 g corn steep solids, 2 g dextrose, 30 g sucrose, 2 g NaNO3, 1 g K2HPO4·3H2O, 0.5 g MgSO4·7H2O, 0.2 g KCl, 0.1 g FeSO4·7H2O, 15 g agar [pH 5.5]) (67). The cultures were incubated for 7 and 20 days at 30°C in the dark. Cultures were imaged with a Cybershot DSC-W120 camera (Sony, New York, NY, USA) before and after ethanol wash, performed to improve the visualization of sclerotia. Micrographs were taken with a Leica MZ75 dissecting microscope attached to a DC50LP camera (Leica Microsystems, Inc., Buffalo Grove, IL, USA).
(vi) Aflatoxin B1 production. In order to assess whether heterologous complementation restores wild-type levels of AFB1 production, the A. flavus wild-type and ΔrtfA, ΔrtfA::An rtfA, and ΔrtfA::Sc rtf1 strains were point-inoculated on YGT medium and incubated for 7 days in the dark at 30°C. Three 16-mm-diameter cores were taken approximately 1 cm from the center of the colony. AFB1 was extracted using 5 ml of chloroform. Extracts were allowed to evaporate and were resuspended in 250 μl of chloroform. Samples were separated using thin-layer chromatography (TLC) on a silica-precoated Polygram Sil G/UV254 TLC plate (Macherey-Nagel, Bethlehem, PA, USA) and chloroform-acetone (85:15 [vol/vol]) as a solvent system. The TLC plate was allowed to air-dry prior to being sprayed with a 12.5% AlCl3 ethanol solution. The TLC plate was then baked at 80°C for 10 min and photographed under UV light (375 nm). Aflatoxin standard was purchased from Sigma-Aldrich (St. Louis, MO, USA). Densitometry of the AF bands in the TLC plates was carried out using the Gelquant.NET software.
Pathogenicity studies. (i) Seed infection assay.
(a) Peanut seed inoculations. The NC94022 Virginia peanut line, kindly provided to us by Baozhu Guo (U.S. Department of Agriculture, Tifton, GA), was utilized in this experiment. The peanut infection experiments were carried out following the procedures described by Zhuang et al. (68), with minor modifications. Briefly, all seeds were shelled, and the embryos were removed and weighed out to approximately 0.25 g to 0.35 g per cotyledon. Each cotyledon was surface-sterilized by being submerged in a 10% Clorox bleach solution for 1 min and then rinsed in sterile double-distilled water (ddH2O) twice to remove the bleach solution. Ten viable cotyledons were then dried and placed in each petri dish. The cotyledons were inoculated on the adaxial surface with 50 µl of conidial suspension (∼105 spores). Cultures were incubated for 5 days at 30°C in the light.
(b) Quantification of conidial production. Groups of four cotyledons infected with each strain were placed in 1.7-ml Eppendorf tubes containing 1 ml of sterile ddH2O. The tubes were vortexed for 1 min. Spores were quantified with a hemocytometer (Hausser Scientific, Horsham, PA) and an Eclipse E-400 bright-field microscope (Nikon, Inc., Melville, NY, USA). The experiment was carried out in triplicate.
(c) AFB1 analysis. Groups of two peanut cotyledons infected with each fungal strain were ground in liquid nitrogen and then added to 12.5 ml of sterile ddH2O in a 50-ml beaker containing 6.25 ml of acetone. The tubes were placed on a rotary platform for 1 h. Each sample was filtered through Whatman paper and collected in another 50-ml Falcon tube, and 17.25 ml of methylene chloride was added. The tubes were inverted 3 times and centrifuged at 3,250 × g for 5 min to separate the organic layers. The bottom organic layer was filtered through granulated sodium sulfate to absorb excess water. Filtrates were evaporated overnight and then resuspended in 2 ml of methylene chloride. The samples were transferred to another Falcon tube, evaporated, resuspended in 300 µl of acetone, and transferred to 1.7-ml Eppendorf tubes. The extracts were again allowed to evaporate in the Eppendorf tubes and were resuspended in 100 μl of acetone. Twenty-five microliters of each extract was separated by TLC, as described above.
(d) Quantification of ergosterol. After incubation at 30°C in the light for 5 days, 2 peanut cotyledons were ground in liquid nitrogen and extracted with a 4-ml solution of chloroform-methanol (2:1 [vol/vol]) overnight at room temperature. Extracts were filtered through sterile Miracloth (Calbiochem, San Diego, CA, USA) into 50-ml beakers. The extracts were allowed to evaporate and then resuspended in 3 ml of the extraction solution. One milliliter of each sample was filtered through a 0.2-μm filter and placed into a 1-ml vial for high-performance liquid chromatography (HPLC) analysis. Twenty-five microliters of each sample was injected into a 1525 HPLC system (Waters, Milford, MA, USA) equipped with a binary pump and a Waters 717 autosampler. HPLC separation occurred at 50°C on a Phenomenex C18 4.6 by 25 mm, 5-μm analytical column equipped with a column guard. With a 2487 dual λ absorbance detector (Waters), UV detection occurred at 282 nm. Samples were quantified using 100% HPLC-grade methanol at a flow rate of 1.0 ml/min. The peaks of samples were then compared to a standard curve of HPLC-grade ergosterol standard (Sigma-Aldrich, St. Louis, MO, USA) to determine the concentration of ergosterol in each sample.
(ii) Galleria mellonella infection model. Ten microliters of a spore suspension of the wild-type, ΔrtfA mutant, and ΔrtfA-com strains in a 1× phosphate-buffered saline (PBS) buffer at a concentration of 1.0 × 104 spores/ml was used to inoculate the G. mellonella larvae, using a technique previously described (45), with minor modifications. Briefly, larvae that were healthy and lacking gray/black markings were selected for the procedure. Approximately 30 larvae were infected with each strain. Thirty larvae infected with 10 μl of a 1× PBS buffer without spores and 10 noninjected larvae served as controls for the experiment. Survival rates were monitored 24 h postinfection. A pairwise comparison statistical analysis was carried out using the SPSS software.
Adhesion study. The ability of A. flavus strains to adhere to surfaces was assessed as previously described (45). One hundred thirty microliters of spore suspension (approximately 7.7 × 104 spores) from each A. flavus strain was added to the wells of a sterile 96-well polystyrene plate, using 32 replicates per strain. The cultures were grown for 24, 48 and 72 h at 30°C in the dark. After incubation, the supernatant and the fungal mycelial mat at the surface were removed. The mycelium adhered to the 96-well wall was washed 3 times with sterile ddH2O. Then, 130 μl of an aqueous 0.01% crystal violet solution was added to each well and allowed to incubate for 20 min. Each well was then washed with 130 μl of sterile ddH2O and allowed to dry. To destain the samples, 130 μl of a 30% acetic acid solution was added to each well, and after 1 min, the plates were imaged with a Cybershot DSC-W120 camera (Sony, New York, NY, USA), and adhesion capacity was estimated by the absorbance at 560 nm using an Epoch spectrophotometer (Biotek, Winooski, VT, USA).
Enzymatic assays.
(i) Protease activity. To assess whether rtfA plays a role in regulating protease activity in A. flavus, strains (106 spores/ml) were inoculated into 500 ml of liquid PMS broth [per liter, 50 g peptone, 3 g (NH4)2SO4, 10 g K2HPO4, 2 g MgSO4·7 H2O, 1 ml of trace elements (pH 5.2)] and incubated at 37°C at 250 rpm for 16 h. Approximately 1 g of mycelium was collected by filtering through Miracloth (Calbiochem, San Diego, CA, USA) and washed with water before being transferred into 25 ml of liquid 0.01% gut microbiota medium (GMM) containing 8 mg/ml of bovine serum albumin (BSA) and incubated at 250 rpm at 30°C to induce protease activity. Fungal supernatants were collected after 24, 48, and 72 h of incubation and filtered through 0.2 μm low-protein-binding filters. Protease activity was measured by an azocasein assay (69) with some minor modifications, as described by Duran et al. (70). Absorbance at 436 nm was read using an Epoch spectrophotometer (Biotek, Winooski, VT, USA).
(ii) Lipase activity. To test a possible role of rtfA in lipase activity, a method utilized by Amaike et al. (71) was used. Briefly, 100 μl of water containing approximately 105 spores of each strain was spread on tributyrin agar plates (per liter, 3 g yeast extract, 5 g peptone, 10 ml tributyrin, 10 g agar [pH 7.5]). Six replicates per strain were used. The inoculated cultures were incubated at 30°C in the dark. Zones of degradation were measured after 72, 96, and 120 h of incubation.
(iii) Amylase assay. To determine if rtfA is necessary for normal amylase activity in A. flavus, a procedure previously described by Duran et al. (70) was utilized, with minor modifications. Briefly, the wild-type, ΔrtfA mutant, and ΔrtfA-com strains (106 spores/ml) were inoculated in a 1-liter flask containing 500 ml of liquid PMS broth. The cultures were incubated at 37°C for 24 h at 250 rpm. Following incubation, mycelia were washed three times with sterile ddH2O, and approximately 1 g of mycelia was transferred into a 125-ml flask containing 25 ml of an amylase-inducing medium (GMM, with 1% starch as the carbon source instead of glucose). The cultures were incubated at 30°C at 250 rpm. Amylase activity was examined after 24, 48, and 72 h of incubation. One hundred microliters of fungal supernatant was mixed with 100 μl of a 0.5% maltoheptose solution and incubated at 40°C for 18 h. The reaction was stopped by heating the mixture at 100°C for 5 min. Approximately 5 μl of the reaction solution was loaded onto a silica-precoated Polygram Sil G/UV254 TLC plate (Macherey-Nagel, Bethlehem, PA, USA) and compared to 5 μl of glucose, maltose, and maltotriose standards (1 mg/ml; Sigma-Aldrich, St. Louis, MO, USA). The TLC plate was then developed in an isopropanol-water-ammonium hydroxide (70:30:10 [vol/vol]) solvent system, sprayed with 30% sulfuric acid, and dried before being charred at 100°C for 5 min. The plates were then imaged with a Cybershot DSC-W120 camera (Sony, New York, NY, USA).
Cell wall analysis.
(i) Determination of cell wall integrity defects. To analyze whether the absence of rtfA results in a loss of integrity of the A. flavus cell wall, the strains were point-inoculated onto YGT medium containing 0.1% sodium dodecyl sulfate (SDS), a cell wall-disrupting compound. Cultures were incubated at 30°C in the dark for 72 h with two replicates per strain. Colony diameters were measured in millimeters.
(ii) Analysis of cell wall composition. (a) Sample preparation. Analysis of cell wall composition was performed to evaluate whether rtfA affects the biosynthesis of cell wall components, specifically mannoprotein, glucan, and chitin, using methods previously described (72). Briefly, the A. flavus strains (106 spores/ml) were inoculated into 50 ml of liquid YGT medium and incubated at 37°C for 42 h at 250 rpm in a rotary shaker. Mycelia were harvested using Miracloth (Calbiochem, San Diego, CA, USA) and washed three times with sterile ddH2O. The mycelia were frozen in liquid nitrogen and stored at −20°C. Prior to the analysis, 100 mg of pulverized mycelia was treated with 1 ml of a cell wall buffer (2% SDS in 50 mM Tris-HCl buffer supplemented with 100 mM Na-EDTA, 40 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and boiled for 15 min to remove any unbound cell wall proteins and water-soluble sugars. After boiling, the buffer was removed and the mycelia washed 3 times with sterile water. The samples were then lyophilized overnight. Approximately 12 mg of lyophilized mycelia for each strain was used for the analysis, with 5 replicates per strain. The samples were treated with 3% NaOH at 75°C for 1 h and centrifuged at 15,000 × g for 15 min. The supernatant was collected and used to quantify mannoprotein and soluble glucan content. The pellet was further digested with 96% formic acid for 4 h at 100°C. After digestion, the formic acid was evaporated, the residue was resuspended in 1 ml of sterile water, and chitin and insoluble glucan content was quantified.
(b) Mannoprotein content. Mannoprotein content was assessed by the bicinchoninic acid (BCA) method described by de Groot et al. (73). Briefly, 10 μl of each sample was mixed with 200 μl of BCA protein assay reagent A (Fisher Scientific, Waltham, MA, USA), and then BCA protein assay reagent B (Fisher Scientific) was added to the mixture at 1:50 (vol/vol) of the total volume. The samples were incubated at 37°C for 30 min, and the absorbance at 560 nm was measured using an Epoch spectrophotometer (Biotek, Winooski, VT, USA). This experiment was carried out with 5 replicates used per strain.
(c) Glucan content. Glucan content was assessed by utilizing the phenol-sulfuric acid method described by Dubois et al. (74). Approximately 200 μl of supernatant (to measure soluble glucan) and 200 μl of the residue (to measure insoluble glucan) solubilized in 1 ml of sterile water were added to 100 μl of a fresh aqueous 5% phenol solution. Then, 500 μl of concentrated sulfuric acid was added to each sample and immediately mixed. The color development was allowed to proceed at room temperature for approximately 30 min. Glucan content (soluble and insoluble) was assessed by measuring glucose levels at an absorbance of 490 nm using an Epoch spectrophotometer (Biotek, Winooski, VT, USA). This experiment was carried out with 5 replicates used per strain.
(d) Chitin content. Chitin concentrations were measured by the method described by Lee et al. (75). After digestion with formic acid, evaporation, and resuspension in 1 ml of water, 100 μl of the sample was mixed with 100 μl of solution A (1.5 M Na2CO3 in 4% [wt/vol] acetylacetone). The reaction mixture was allowed to incubate at 100°C for approximately 20 min. After cooling at room temperature, 700 μl of 95% ethanol (EtOH) and 100 μl of solution B (1.6 g p-dimethylaminobenzaldehyde in 30 ml of concentrated HCl and 30 ml of 95% EtOH) were added to each sample and allowed to incubate at room temperature for 1 h. Chitin content was determined by measuring N-acetyl-d-glucosamine levels at an absorbance of 520 nm using an Epoch spectrophotometer (Biotek, Winooski, VT, USA). This experiment was carried out with 5 replicates per strain.
Oxidative stress sensitivity assessment.
To evaluate whether rtfA is important for resistance to oxidative stress in A. flavus, we used the method described by Baidya et al. (76). Briefly, the strains were point-inoculated on 3 ml of YGT solid medium supplemented with various concentrations of menadione, a compound used to induce the production of reactive oxygen species (ROS) (77). The menadione concentrations applied in this assay were 0, 0.1, 0.2, and 0.4 mM. Cultures were incubated for 72 h at 30°C in the dark before being imaged with a with a Cybershot DSC-W120 camera (Sony, New York, NY, USA). The experiment was repeated 3 times with similar results.
Metabolomics analysis.
(i) Sample collection and extraction. The A. flavus wild-type, ΔrtfA mutant, and ΔrtfA-com strains were inoculated in 25 ml of liquid YGT medium at a concentration of 106 spores/ml. Stationary liquid cultures were incubated for 5 days at 30°C in the dark. Culture supernatants were collected, and A. flavus secondary metabolites were extracted with chloroform using a 1:1 ratio. The chloroform layer was collected and allowed to evaporate overnight in a 50-ml beaker.
(ii) LC-MS. Sample analysis was performed using an HPLC system coupled to an LTQ Orbitrap XL high-resolution mass spectrometer (Thermo Fisher Scientific, Les Ulis, France). Extracts were solubilized in 500 µl of water-acetonitrile (vol/vol), and 10 μl of this solution was injected into a reversed-phase (150 mm by 2.0 mm) 5-μm Luna C18 column (Phenomenex, Torrance, CA, USA) operated at a flow rate of 0.2 ml/min. A gradient program was performed with 0.05% formic acid (phase A) and 100% acetonitrile (phase B) with the following elution gradient: 0 min, 20% B; 30 min, 50% B; 35 to 45 min, 90% B; and 50 to 60 min, 20% B. High-resolution mass spectrometry (HRMS) acquisitions were achieved with electrospray ionization (ESI) in the positive and negative modes, as follows: spray voltage of +5.5 kV, capillary temperature of 350°C, sheath gas (N2) flow rate of 30 arbitrary units (au), and auxiliary gas (N2) flow rate of 10 au in the positive mode; and spray voltage of −3.7 kV, capillary temperature of 350°C, sheath gas (N2) flow rate of 30 au, and auxiliary gas (N2) flow rate of 10 au in the negative mode. Full MS spectra were acquired at a resolution of 60,000 with a range of mass-to-charge ratio (m/z) set to 100 to 800, while the MS/MS spectra were acquired at low resolution. The identity of fungal products was confirmed by comparison either with HPLC-MS2 analysis of a standard compound or on the basis of results obtained in Carvajal-Campos et al. (78).
Statistical analysis.
Statistical analysis was carried out for all quantitative data in this study. Analysis of variance (ANOVA) in conjunction with Tukey’s post hoc test was carried out using the statistical software program R version x64 3.3.0.
Data availability.
The accession numbers corresponding to all sequences used in this study are listed in Table S1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Baozhu Guo (USDA, Tifton, GA) for kindly providing the NC94022 Virginia peanut line used in this study.
This work was supported by USDA grant 58-6435-4-015 and the Department of Biological Sciences at Northern Illinois University.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02446-18.
REFERENCES
- 1.Bhatnagar D, Brown R, Ehrlich K, Cleveland TE. 2002. Mycotoxins contaminating cereal grain crops: their occurrence and toxicity. In Khachatourians GG, Aora DK (ed), Applied mycology and biotechnology, vol 2. Agricultural and food production. Elsevier BV, Amsterdam, Netherlands. [Google Scholar]
- 2.Cary JW, Linz JE, Bhatnagar D (ed). 2000. Microbial foodborne diseases: mechanisms of pathogenesis and toxin synthesis, p 317–361. Technomic Publishing Co., Lancaster, PA. [Google Scholar]
- 3.Sweeny MJ, Dobson AD. 1999. Molecular biology of mycotoxin biosynthesis. FEMS Microbiol Lett 175:149–163. [DOI] [PubMed] [Google Scholar]
- 4.Trail F, Mahanti N, Linz JE. 1995. Molecular biology of aflatoxin biosynthesis. Microbiology 141:755–765. doi: 10.1099/13500872-141-4-755. [DOI] [PubMed] [Google Scholar]
- 5.Hsieh DP. 1988. Potential human health hazards of mycotoxins, p 69–80. In Natori SHK, Ueno Y (ed), Mycotoxins and phytotoxins. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 6.Turner PC, Moore SE, Hall AJ, Prentice AM, Wild CP. 2003. Modification of immune function through exposure to dietary aflatoxin in Gambian children. Environ Health Perspect 111:217–220. doi: 10.1289/ehp.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Probst C, Njapau H, Cotty PJ. 2007. Outbreak of an acute aflatoxicosis in Kenya in 2004: identification of the causative agent. Appl Environ Microbiol 73:2762–2764. doi: 10.1128/AEM.02370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgianna DR, Fedorova ND, Burroughs JL, Dolezal AL, Bok JW, Horowitz-Brown S, Woloshuk CP, Yu J, Keller NP, Payne GA. 2010. Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol Plant Pathol 11:213–222. doi: 10.1111/j.1364-3703.2009.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. 2010. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terabayashi Y, Sano M, Yamane N, Marui J, Tamano K, Sagara J, Dohmoto M, Oda K, Ohshima E, Tachibana K, Higa Y, Ohashi S, Koike H, Machida M. 2010. Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet Biol 47:953–961. doi: 10.1016/j.fgb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Riley RT, Goeger DE. 1992. Cyclopiazonic acid: speculations on its function in fungi, p 385–402. In Bhatnagar D, Lillehoj EB, Arora DK (ed), Handbook of applied mycology. Mycotoxins in ecological systems. Marcel Dekker, New York, NY. [Google Scholar]
- 12.Valdes JJ, Cameron JE, Cole RJ. 1985. Aflatrem: a tremorgenic mycotoxin with acute neurotoxic effects. Environ Health Perspect 62:459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao I, Peter AB, Baur R, Sigel E. 1988. The tremorigen aflatrem is a positive allosteric modulator of γ-aminobutyric acid A receptor channel expressed in Xenopus oocytes. Mol Pharmacol 35:319–323. [PubMed] [Google Scholar]
- 14.Zhang C, Jin L, Mondie B, Mitchell SS, Castelhano AL, Cai W, Bergenhem N. 2003. Leporin B: a novel hexokinase II gene inducing agent from an unidentified fungus. Bioorg Med Chem Lett 13:1433–1435. doi: 10.1016/S0960-894X(03)00153-7. [DOI] [PubMed] [Google Scholar]
- 15.Gloer JB, TePaske MR, Sima JS, Wicklow DT, Dowd PF. 1988. Antiinsectan aflavinine derivatives from the sclerotia of Aspergillus flavus. J Org Chem 53:5457–5460. doi: 10.1021/jo00258a011. [DOI] [Google Scholar]
- 16.Kishimoto S, Sato M, Tsunematsu Y, Watanabe K. 2016. Evaluation of biosynthetic pathway and engineered biosynthesis of alkaloids. Molecules 21:1–19. doi: 10.3390/molecules21081078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monti F, Ripamonti F, Hawser SP, Islam K. 1999. Aspirochlorine: a highly selective and potent inhibitor of fungal protein synthesis. J Antibiot (Tokyo) 52:311–318. [DOI] [PubMed] [Google Scholar]
- 18.Wu F, Guvlu H. 2012. Aflatoxin regulations in a network of global maize trade. PLoS One 7:e435151. doi: 10.1371/journal.pone.0045151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu F. 2015. Global impacts of aflatoxin in maize: trade and human health. World Mycotoxin J 8:137–142. doi: 10.3920/WMJ2014.1737. [DOI] [Google Scholar]
- 20.Mitchell NJ, Bowers E, Hurburgh C, Wu F. 2016. Potential economic losses to the USA corn industry from aflatoxin contamination. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 33:540–550. doi: 10.1080/19440049.2016.1138545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosquera J, Warn PA, Morrissey J, Moore CB, Gil-Lamaignere C, Denning DW. 2001. Susceptibility testing of Aspergillus flavus: inoculum dependence with itraconazole and lack of correlation between susceptibility to amphotericin B in vitro and outcome in vivo. Antimicrob Agents Chemother 45:1456–1462. doi: 10.1128/AAC.45.5.1456-1462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamai Y, Harasaki T, Fukuoka T, Ohya S, Uchida K, Yamaguchi H, Kuwahara S. 2002. In vitro and in vivo activities of CS-758 (R-120758), a new triazole antifungal agent. Antimicrob Agents Chemother 46:367–370. doi: 10.1128/AAC.46.2.367-370.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW. 2007. Aspergillus flavus: human pathogen, allergen, and mycotoxin producer. Microbiology 153:1677–1692. doi: 10.1099/mic.0.2007/007641-0. [DOI] [PubMed] [Google Scholar]
- 24.Ford S, Friedman L. 1967. Experimental study of the pathogenicity of aspergilli for mice. J Bacteriol 94:928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaliamurthy J, Geraldine JP, Thomas PA. 2003. Disseminated aspergillosis due to Aspergillus flavus in an experimental model: efficacy of azole therapy. Mycoses 46:174–182. [DOI] [PubMed] [Google Scholar]
- 26.Chakrabarti A, Gupta V, Biswas G, Kumar B, Sakhuja VK. 1998. Primary cutaneous aspergillosis: our experience in 10 years. J Infect 37:24–27. [DOI] [PubMed] [Google Scholar]
- 27.Hussain S, Salahuddin N, Ahmad I, Salahuddin I, Jooma R. 1995. Rhinocerebral invasive mycosis: occurrence in immunocompetent individuals. Eur J Radiol 20:151–155. [DOI] [PubMed] [Google Scholar]
- 28.Iwen PC, Rupp ME, Hinrichs SH. 1997. Invasive mold sinusitis: 17 cases in immunocompromised patients and review of the literature. Clin Infect Dis 24:1178–1184. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy CA, Adams GL, Neglia JP, Giebink GS. 1997. Impact of surgical treatment on paranasal fungal infections in bone marrow transplant patients. Otolaryngol Head Neck Surg 116:610–616. [DOI] [PubMed] [Google Scholar]
- 30.Panda NK, Sharma SC, Chakrabartu A, Mann SBS. 1998. Paranasal sinus mycoses in north India. Mycoses 41:281–286. [DOI] [PubMed] [Google Scholar]
- 31.Khairallah SH, Byrne KA, Tabbara KF. 1992. Fungal keratitis in Saudi Arabia. Doc Ophthalmol 79:269–276. [DOI] [PubMed] [Google Scholar]
- 32.Roilides E, Simitsopoulou M, Katragkou A, Walsh TJ. 2015. How biofilms evade host defenses. Microbiol Spectr 3:MB-0012-2014. doi: 10.1128/microbiolspec.MB-0012-2014. [DOI] [PubMed] [Google Scholar]
- 33.Dolezal AL, Obrian GR, Nielsen DM, Woloshuk CP, Boston RS, Payne GA. 2013. Localization, morphology and transcriptional profile of Aspergillus flavus during seed colonization. Mol Plant Pathol 14:898–909. doi: 10.1111/mpp.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellon JE, Cotty PJ, Dowd MK. 2007. Aspergillus flavus hydrolases: their roles in pathogenesis and substrate utilization. Appl Microbiol Biotechnol 77:497–504. doi: 10.1007/s00253-007-1201-8. [DOI] [PubMed] [Google Scholar]
- 35.Latge JP. 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol 66:279–290. doi: 10.1111/j.1365-2958.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- 36.Selvig K, Alspaugh JA. 2011. pH response pathways in fungi: adapting to host-derived and environmental signals. Mycobiology 39:249–256. doi: 10.5941/MYCO.2011.39.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 38.Jaehning JA. 2010. The Paf 1 complex: platform or player in RNA polymerase II transcription? Biochem Biophys Acta 1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller CL, Jaehning JA. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol 22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng HH, Dole S, Struhl K. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquination of histone H2B. J Biol Chem 278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- 41.Stolinski LA, Eisenmann DM, Arndt KM. 1997. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol Cell Biol 17:4490–5000. doi: 10.1128/MCB.17.8.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun ZW, Allis CD. 2002. Ubiquination of H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 43.Warner MH, Roinick KL, Arndt KM. 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol 27:6103–6115. doi: 10.1128/MCB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramamoorthy V, Shantappa S, Dhingra S, Calvo AM. 2012. veA-dependent RNA-pol II transcription elongation factor-like protein, RtfA, is associated with secondary metabolism and morphological development in Aspergillus nidulans. Mol Microbiol 85:795–814. doi: 10.1111/j.1365-2958.2012.08142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyers RR, Smith TD, Elsawa SF, Puel O, Tadrist S, Calvo AM. 2017. rtfA controls development, secondary metabolism, and virulence in Aspergillus fumigatus. PLoS One 12:e0176702. doi: 10.1371/journal.pone.0176702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohmar JM, Harris-Coward PY, Cary JW, Dhingra S, Calvo AM. 2016. rtfA, a putative RNA-Pol II transcription elongation factor gene, is necessary for normal morphological and chemical development in Aspergillus flavus. Appl Microbiol Biotechnol 100:5029–5041. doi: 10.1007/s00253-016-7418-7. [DOI] [PubMed] [Google Scholar]
- 47.de Jong RN, Truffault V, Diercks T, Ab E, Daniels MA, Kaptein R, Folkers GE. 2008. Structure and DNA binding of the human Rtf1 Plus3 domain. Structure 16:149–159. doi: 10.1016/j.str.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 48.Ramage G, Rajendran R, Gutierrez-Correa M, Jones B, Williams C. 2011. Aspergillus biofilms: clinical and industrial significance. FEMS Microbiol Lett 324:89–97. doi: 10.1111/j.1574-6968.2011.02381.x. [DOI] [PubMed] [Google Scholar]
- 49.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 50.de Groot PWJ, Ruiz C, Vázquez de Aldana CR, Duenas E, Cid VJ, Del Rey F, Rodríquez-Peña JM, Pérez P, Andel A, Caubín J, Arroyo J, García JC, Gil C, Molina M, García LJ, Nombela C, Klis FM. 2001. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp Funct Genomics 2:124–142. doi: 10.1002/cfg.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujiwara M, Ichinomiya I, Motoyama T, Horiuchi H, Ohta A, Takagi M. 2000. Evidence that the Aspergillus nidulans class I and class II chitin synthase genes, chsC and chsA, share critical roles in hyphal wall integrity and conidiophore development. J Biochem 127:359–366. [DOI] [PubMed] [Google Scholar]
- 52.Forseth RR, Amaike S, Schwenk D, Affeldt KJ, Hoffmeister D, Schroeder FC, Keller NP. 2013. Homologous NRPS-like gene clusters mediate redundant small-molecule biosynthesis in Aspergillus flavus. Angew Chem Int Ed 52:1590–1594. doi: 10.1002/anie.201207456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh S, Zhang H, Ludwig P, van Nocker S. 2004. A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the arabidopsis FLC/MAF MADS box gene family. Plant Cell 16:2940–2953. doi: 10.1105/tpc.104.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tenney K, Gerber M, Ilvarsonn A, Schneider J, Gause M, Dorsett D, Eissenberg JC, Shilatifard A. 2006. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci U S A 103:11970–11974. doi: 10.1073/pnas.0603620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mueller JE, Canze M, Bryk M. 2006. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics 173:557–567. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 58.Shilatifard A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 2006:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 59.Lieb JD, Clarke ND. 2005. Control of transcription through intragenic patterns of nucleosome composition. Cell 123:1187–1190. doi: 10.1016/j.cell.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 60.Damveld RA, Franken A, Arentshorst M, Punt PJ, Klis FM, van den Hondel CAMJJ, Arthur FJ, Ram AFJ. 2008. A novel screening method for cell wall mutants in Aspergillus niger identifies UDP-galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics 178:873–881. doi: 10.1534/genetics.107.073148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mack I, Hector A, Ballbach M, Kohlhäufl J, Fuchs KJ, Weber A, Mall MA, Hartl D. 2015. The role of chitin, chitinases, and chitinase-like proteins in pediatric lung diseases. Mol Cell Pediatr 2:3. doi: 10.1186/s40348-015-0014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown GD, Gordon S. 2003. Fungal β-glucans and mammalian immunity. Immunity 19:311–315. doi: 10.1016/S1074-7613(03)00233-4. [DOI] [PubMed] [Google Scholar]
- 63.Dagenais TR, Keller NP. 2009. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev 22:447–465. doi: 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilbert MK, Majumdar R, Rajasekaran K, Chen ZY, Wei Q, Sickler CM, Lebar MD, Cary JW, Frame BR, Wang K. 2018. RNA interference-based silencing of the alpha-amylase (amy1) gene in Aspergillus flavus decreases fungal growth and aflatoxin production in maize kernels. Planta 247:1465–1473. doi: 10.1007/s00425-018-2875-0. [DOI] [PubMed] [Google Scholar]
- 65.Hill TW, Kafer E. 2001. Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. Fungal Genet Rep 48:20–21. doi: 10.4148/1941-4765.1173. [DOI] [Google Scholar]
- 66.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. 2007. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc 1:3111–3121. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 67.Chang PK, Scharfenstein LL, Mack B, Ehrlich KC. 2012. The deletion of the Aspergillus flavus orthologue of A. nidulans fluG reduces conidiation and promotes the production of sclerotia but does not abolish aflatoxin biosynthesis. Appl Environ Microbiol 78:7557–7563. doi: 10.1128/AEM.01241-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhuang Z, Lohmar JM, Satterlee T, Cary JW, Calvo AM. 2016. The master transcription factor mtfA governs aflatoxin production, morphological development and pathogenicity in the fungus Aspergillus flavus. Toxins (Basel) 8:29. doi: 10.3390/toxins8010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reichard U, Monod M, Odds F, Ruchel R. 1997. Virulence of an aspergillopepsin-deficient mutant of Aspergillus fumigatus and evidence for another aspartic proteinase linked to the fungal cell wall. J Med Vet Mycol 35:189–196. [DOI] [PubMed] [Google Scholar]
- 70.Duran RM, Gregerson S, Smith TD, Behtariya PJ, Cary JW, Harris-Coward PY, Mattison CP, Grimm C, Calvo AM. 2014. The role of Aspergillus flavus veA in the production of extracellular proteins during the growth on starch substrates. Appl Microbiol Biotechnol 98:5081–5094. doi: 10.1007/s00253-014-5598-6. [DOI] [PubMed] [Google Scholar]
- 71.Amaike S, Affeldt KJ, Yin WB, Franke S, Choithani A, Keller NP. 2013. The bZIP protein MeaB mediates virulence attributes in Aspergillus flavus. PLoS One 8:e74030. doi: 10.1371/journal.pone.0074030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feng X, Ramamoorthy V, Pandit SS, Prieto A, Espeso EA, Calvo AM. 2017. cpsA regulates mycotoxin production, morphogenesis and cell wall biosynthesis in the fungus Aspergillus nidulans. Mol Microbiol 105:1–24. doi: 10.1111/mmi.13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Groot PWJ, Kraneveld EA, Yin QY, Dekker HL, Groß U, Crielaard W, de Koster CG, Bader O, Klis FM, Weig M. 2008. The cell wall of the human pathogen Candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryot Cell 7:1951–1964. doi: 10.1128/EC.00284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. 1956. Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- 75.Lee JI, Yu YM, Rho YM, Park BC, Choi JH, Park HM, Maeng PJ. 2005. Differential expression of the chsE gene encoding a chitin synthase of Aspergillus nidulans in response to developmental status and growth conditions. FEMS Microbiol Lett 249:121–129. doi: 10.1016/j.femsle.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 76.Baidya S, Duran RM, Lohmar JM, Harris-Coward PY, Cary JW, Hong SY, Roze LV, Linz JE, Calvo AM. 2014. VeA is associated with the response to oxidative stress in the aflatoxin producer Aspergillus flavus. Eukaryot Cell 13:1095–1103. doi: 10.1128/EC.00099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Criddle DN, Gillies SS, Baumgartner-Wilson HK, Jaffar M, Chinje EC, Passmore S, Chvanov M, Barrow S, Gerasimenko OV, Tepikin AV, Sutton R, Petersen OH. 2006. Menadione-induced reactive oxygen species generation via redox cycling promotes apoptosis of murine pancreatic acinar cells. J Biol Chem 281:40485–40492. doi: 10.1074/jbc.M607704200. [DOI] [PubMed] [Google Scholar]
- 78.Carvajal-Campos A, Manizan AL, Tadrist S, Akaki DK, Koffi-Nevry R, Moore GG, Fapohunda SO, Bailly S, Montet D, Oswald IP, Lorber S, Brabet C, Puel O. 2017. Aspergillus korhogoensis, a novel aflatoxin producing species from the Côte d’Ivoire. Toxins 9:353. doi: 10.3390/toxins9110353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cary JW, Han Z, Yin Y, Lohmar JM, Shantappa S, Harris-Coward PY, Mack BM, Ehrlich KC, Wei Q, Arroyo-Manzanares N, Uka V, Vanhaecke L, Bhatnagar D, Yu J, Nierman WC, Johns M, Sorensen D, Shen H, De Saeger S, Diana Di Mavungu J, Calvo AM. 2015. Transcriptome analysis of Aspergillus flavus reveals veA-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster. Eukaryot Cell 14:983–997. doi: 10.1128/EC.00092-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers corresponding to all sequences used in this study are listed in Table S1.