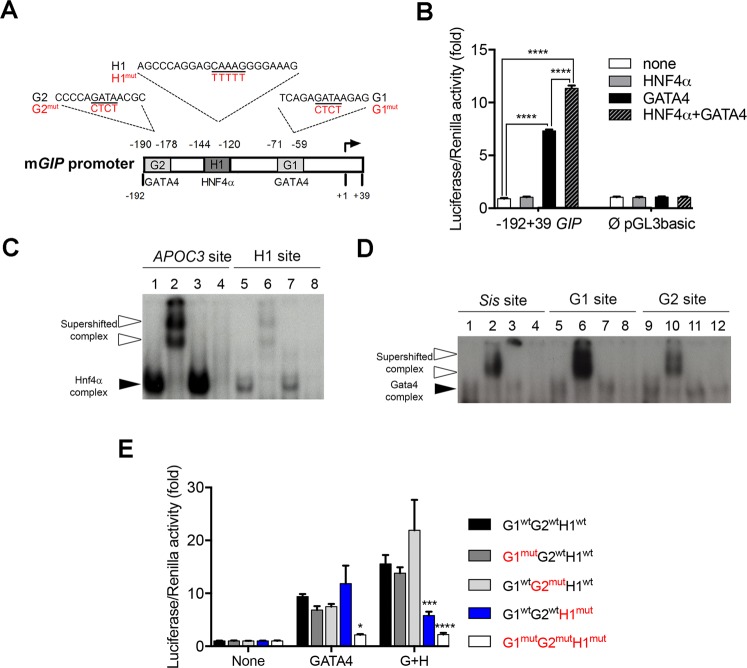
Figure 2.
The mouse Gip promoter is regulated by HNF4α. (A) Schematic representation of the mouse Gip promoter with predicted binding sites for HNF4α and GATA4. Mutations introduced in these sites are indicated in red. (B) Luciferase assay using the −192 +39 Gip-PGL3basic and Empty-pGL3basic constructs with a combination of HNF4α and/or GATA4 expression vectors. Luciferase assays are representative of three independent.experiments and are presented as mean ± SEM; ****P < 0.0001. (C) EMSA analysis with HNF4α or (D) GATA4 nuclear extracts of each potential binding site. Lines 1, 5 and 9 show 32P-labelled probes with nuclear extracts; lines 2, 6 and 10 show 32P-labelled probes with nuclear extracts and antibodies; lines 3, 7 and 11 show 32P-labelled probes with nuclear extracts and irrelevant antibodies; lines 4, 8 and 12 show 32P-labelled probe with nuclear extracts from non-transfected empty cells. White arrowheads point to supershifted complexes while black arrowheads depict retarded complexes. Full length gels of these analyses are provided in Supplementary Fig. 2. (E) Luciferase assay using the −192 +39 Gip-PGL3 wild-type or mutated promoter constructs in combination of HNF4α (H) and/or GATA4 (G) expression vectors. Mutation effects on luciferase activity are evaluated against the wild-type promoter for each condition. Luciferase assays are representative of three independent experiments and are presented as mean ± SEM; *P < 0.05, ***P < 0.001, ****P < 0.0001.