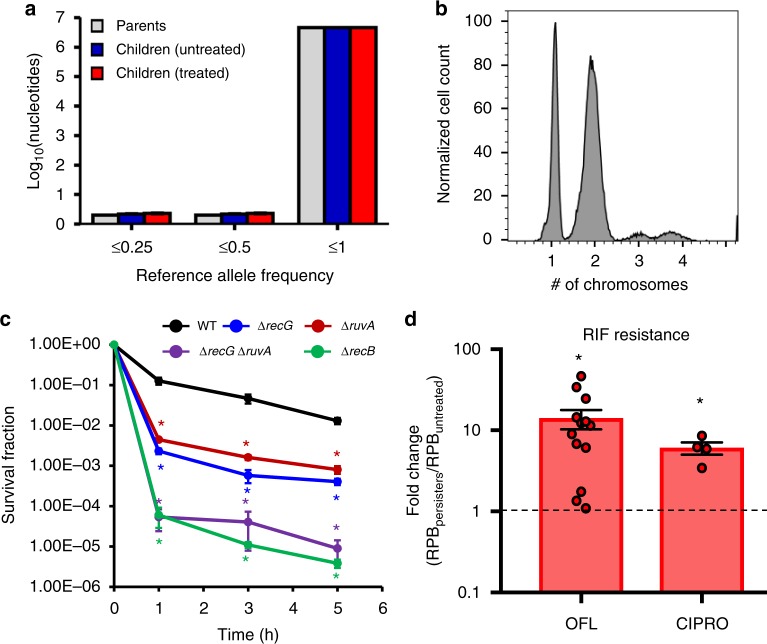
Fig. 5.
Fidelity of DNA repair in OFL persisters. a WGS was performed from DNA extracted from 10 persisters, 10 untreated controls, and 4 parental cultures, and number of nucleotides with reference allele frequencies (RAF) less than or equal to the indicated values are depicted in the histogram. RAF of 1 indicates that all sequencing reads of that nucleotide were of the reference nucleotide, whereas RAF of 0 indicates that all sequence reads had a non-reference nucleotide. Values between 0 and 1 indicate that some reads were of the reference nucleotide. The RAFs at different thresholds were indistinguishable among the persisters, untreated controls, and parental cultures, suggesting that OFL persisters generally repair DNA damage correctly. b Chromosomal content of stationary-phase cultures prior to OFL treatment was determined using PicoGreen stains (n = 3; see Supplementary Fig. 8i for controls used to determine fluorescence intensities corresponding to chromosome number). 30.8 ± 1.6% of cells contained one chromosome, whereas 68.4 ± 1.6% of cells contained two or more chromosomes, indicating that the majority of cells had the chromosomal content for repair by homologous recombination. Histogram shown is representative of three biological replicates. c Mutants defective in homologous recombination, which requires the resolution of Holliday junctions by RuvA and/or RecG, demonstrate reduced survival to OFL treatment in stationary phase (n = 3). d Enhancement in RIF resistance was observed in progeny of CIPRO persisters (n = 4), similar to offspring of OFL persisters, demonstrating that the phenomenon of resistance enhancement following treatment and recovery is shared between distinct fluoroquinolones. Error bars depict S.E.M. For c, * indicates significance between log-transformed survival fractions and those of wild-type (p ≤ 0.05, using two-tailed t-tests with unequal variances). For d, a value of 1 is indicative of equivalent abundances (no change), and * denotes a log-transformed fold-change that is significantly different compared to a log-transformed value of 1 (p ≤ 0.05, using two-tailed t-tests with unequal variance)