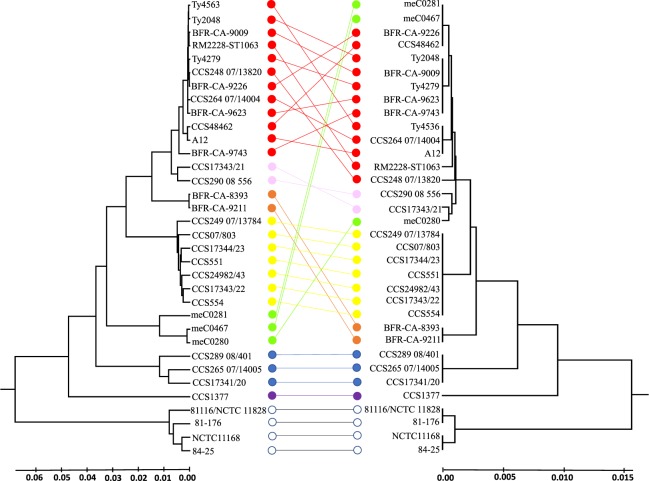
Figure 4.
Comparison of MLST-based and proteotyping-based UPGMA dendrograms. The MLST-based phylogenetic tree (left) as well as the proteotyping-based dendrogram (right) were constructed by UPGMA. The MLST dendrogram resulted from 7 loci, the proteotyping-based dendrogram from the amino acid sequences of 16 identified biomarker ions. The different C. coli clades and sub-clades are represented by different colors. In addition, four C. jejuni isolates have been included in the illustration, which form their own C. jejuni clade. Color codes: clade 1A – red, clade 1B – pink, clade 1C – orange, clade 2 – yellow, clade 3 – blue, isolate CCS1377 – purple, isolates meC0280, mecC0281, and meC0467 – green, C. jejuni isolates – white. Lines connect the corresponding isolates in the different trees. As it can be seen, there are only crossings of connecting lines within one clade (corresponding to one color), whereas different colors (clades) do not cross each other. This demonstrates that proteotyping can be used to distinguish the clades clearly from each other. The only exceptions are the three isolates meC0280, mecC0281, and meC0467 labeled in green. These form their own clade in the MLST-based tree (Supplementary Fig. 1), but in the core genome alignment (Supplementary Fig. 2) they cluster with C. coli clade 1. This means that for isolates of this group the proteotyping-based tree is similar to a core genome alignment, while MLST is less suitable.