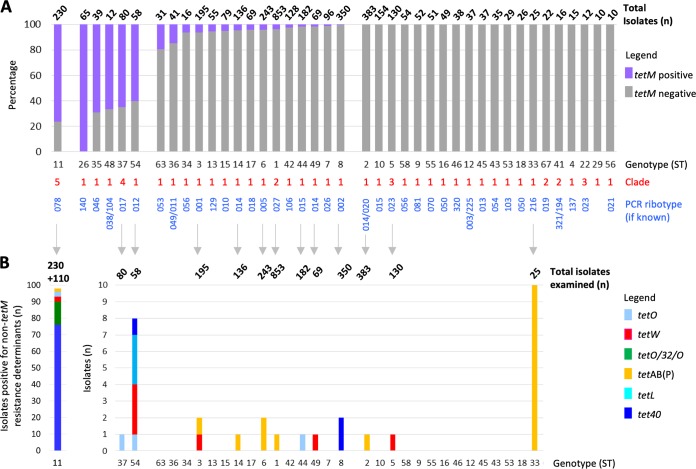
FIG 1.
Prevalence of tetracycline resistance determinants in RT078 and other clinically relevant C. difficile genotypes. (A) Proportion (percentage) of each clinically important genotype that was positive for the ribosomal protection protein (RPP) gene tetM. Data are shown for genotypes having 10 genomes or more, from isolate collections representing Oxfordshire (EIA positives and negatives, infant and farm) and Leeds, North America, and Europe (Optimer clinical trial) (8, 29, 30, 33, 34). The total number of isolates of each genotype is shown above the bar. Clades are defined as described in reference 69. (B) Numbers of genomes in the collections described above which contained additional non-tetM tetracycline resistance determinants. For the ST11(078) genotype, the additional Scottish (n = 110) isolate collection was also included (indicated by “+110” above the bar at the left). Therefore, a total of 340 ST11(078) isolates were examined (the n = 230 described in the panel A legend above plus an additional n = 110 Scottish ST11s), the aim being to illustrate the overall prevalence of “non-tetM” tetracycline resistance determinants within this genotype.