Figure 3.
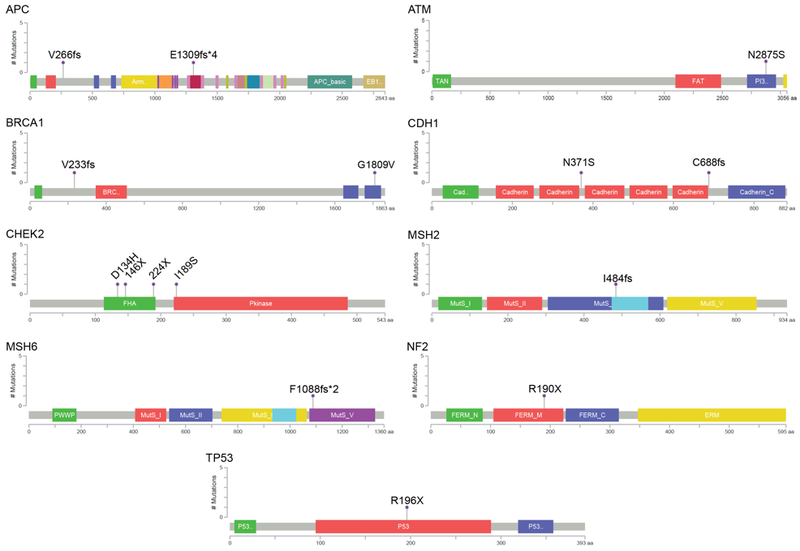
Presumed pathogenic germline variants. The locations of mutations and domains in the encoded protein products (shown by different colors) for the 15 presumed pathogenic variants are shown in lollipop plots. On the graph of each gene, the x axis reflects the number of amino acid residues, and the y axis represents the total number of mutations identified. Frameshift and stop gain mutations were classified in Tier 1, and were found in the following genes (as denoted by chromosomal locations): APC (5q22.2), BRCA1 (17q21.31), CDH1 (16q22.1), CHEK2 (22q12.1), MSH2(2p21-p16.3), MSH6 (2p16.3), NF2 (22q12.2), TP53 (17p13.1). Missense mutations were classified into Tiers 2 and 3, and were found in the following genes (as denoted by chromosomal locations): ATM (11q22.3), BRCA1 (17q21.31), CDH1 (16q22.1), CHEK2 (22q12.1).
