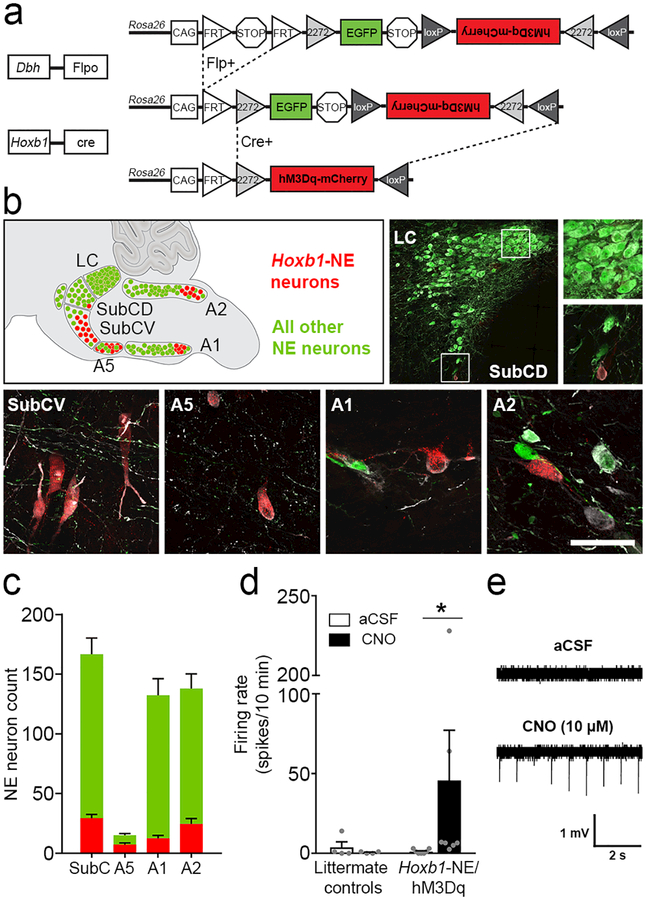
Figure 1. Intersectional chemogenetic strategy to selectively activate Hoxb1-NE neurons.
(a) Schematic diagram of the RC::FL-hM3Dq allele. Recombination by DbhFlpo leads to EGFP expression in NE neurons, and recombination by both Hoxb1Cre and DbhFlpo leads to hM3Dq-mCherry expression in Hoxb1-NE neurons. (b) Approximate positions of noradrenergic nuclei are illustrated on a sagittal schematic diagram of the hindbrain compressed along the mediolateral axis. Immunofluorescent labeling of parasagittal sections from Hoxb1-NE/hM3Dq brains for tyrosine hydroxylase (white), mCherry (red) and EGFP (green) reveals hM3Dq-mCherry-expressing Hoxb1-NE neurons in dorsal and ventral subcoeruleus (SubCD and SubCV), A5, caudal A1, and caudal A2 noradrenergic nuclei, but not in the locus coeruleus. Scale bar: 287 μm (LC), 100 μm (LC and SubCD close-ups), 50 μm (SubCV, A5, A1, A2). (c) Counts of hM3Dq-mCherry-expressing (red bars) and EGFP-expressing NE neurons (green bars) in different nuclei (n=12 mice). (d) Bath application of CNO (10 μM) increases firing rate in hM3Dq-mCherry-expressing neurons (Hoxb1-NE/hM3Dq mice, n=7 cells from 5 mice), but not in EGFP-expressing neurons (littermate control mice, n=4 cells from 2 mice). Data are mean ± SEM. *P<0.05 vs. aCSF, Wilcoxon signed ranks test. (e) Representative traces of cell-attached recordings from hM3Dq-mCherry-expressing neurons before (top) and after (bottom) bath application of CNO (10 μM).