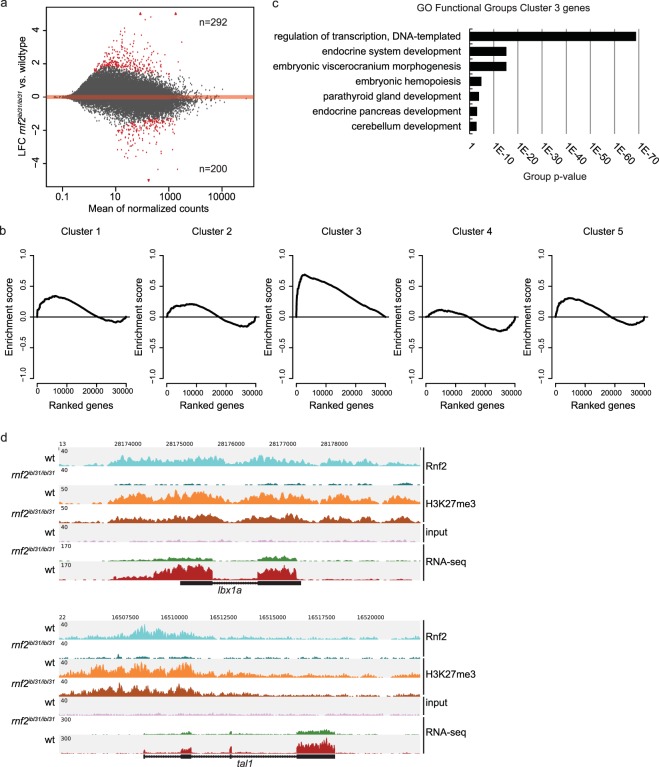
Figure 3.
Loss of Rnf2 is directly associated with upregulation of gene expression. (a) MA-plot of differentially expressed genes between wildtypes and rnf2 mutants at 3 dpf. Significantly differentially expressed genes |Log2FC ≥ 1|; padj < 0.1 are highlighted in red. In total 292 genes are upregulated and 200 genes are downregulated. (b) Gene Set Enrichment Analysis (GSEA) for the genes whose promoters belong to the five clusters defined in Fig. 2a. Cluster 1, 2, 4, and 5 show a non-significant distribution of the genes based on their expression changes upon the rnf2 mutation. Genes belonging to the promoters from cluster 3 are enriched for upregulation upon mutation of rnf2. p-value = < 0.001; NES = 2.04. (c) Gene ontology of biological processes analysis of the 112 genes belonging to cluster 3. (d) ChIP-seq and RNA-seq coverage at the lbx1a gene and the tal1 gene in 3 dpf zebrafish embryos. Light blue: Rnf2 ChIP-seq tracks in wildtypes. Teal: Rnf2 ChIP-seq tracks in rnf2 mutants. Orange: H3K27me3 ChIP-seq track in wildtypes. Brown: H3K27me3 ChIP-seq track in rnf2 mutants. Lilac: wildtype input. Green: RNA-seq track in wildtypes. Red: RNA-seq track in rnf2 mutants.