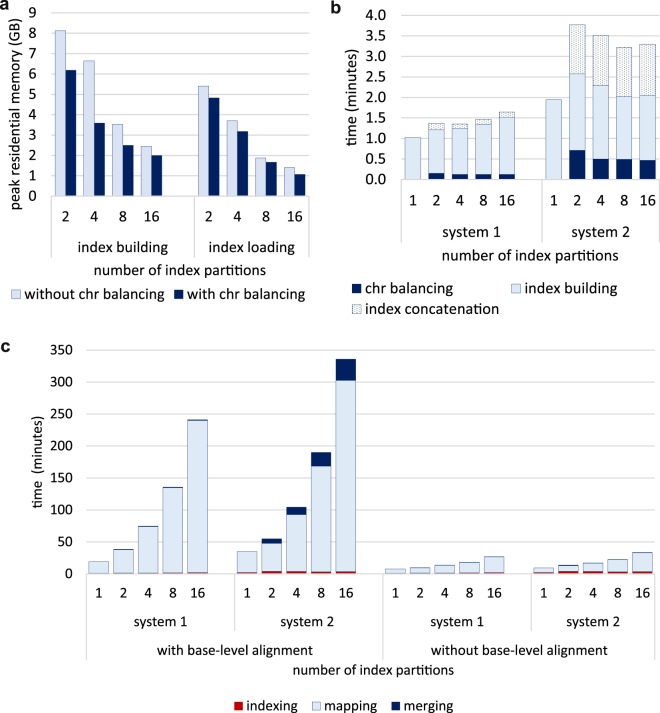
Figure 5.
Peak memory usage and runtime for a partitioned index of the human genome. (a) Peak memory usage in function of the number of partitions for ab initio index generation (left) and loading a pre-built index (right). Dark bars represent memory usage when performing chromosome balancing as described herein, whilst light bars represent the default iterative partition distribution as implemented in Minimap2. (b) Detailed runtime metrics for index building across two computational systems. System 1 is a laptop with flash memory (Intel i7-8750H processor, 16 GB of RAM and Toshiba XG5 series NVMe SSD) while system 2 is a workstation with a mechanical hard disk (Intel i7-6700 processor, 16 GB of RAM and Toshiba DT01ACA series HDD). The total indexing time has been broken down into three steps; chr balancing, index building and index concatenations. Chr balancing includes the overhead for chromosome sorting, partitioning and writing each partition to a separate file. (c) Runtime for base-level alignment (left) and block/locus level mapping (right). System 1 and 2 are as described in (b). Alignment was performed on the NA12878 data (see Materials and methods) with the map-ont pre-set in Minimap2 using 8 threads. Runtime statistics are composed of indexing (index generation including the overhead), mapping (total time for aligning reads to each partition), and merging using the method described herein. Runtimes include file reading and writing.