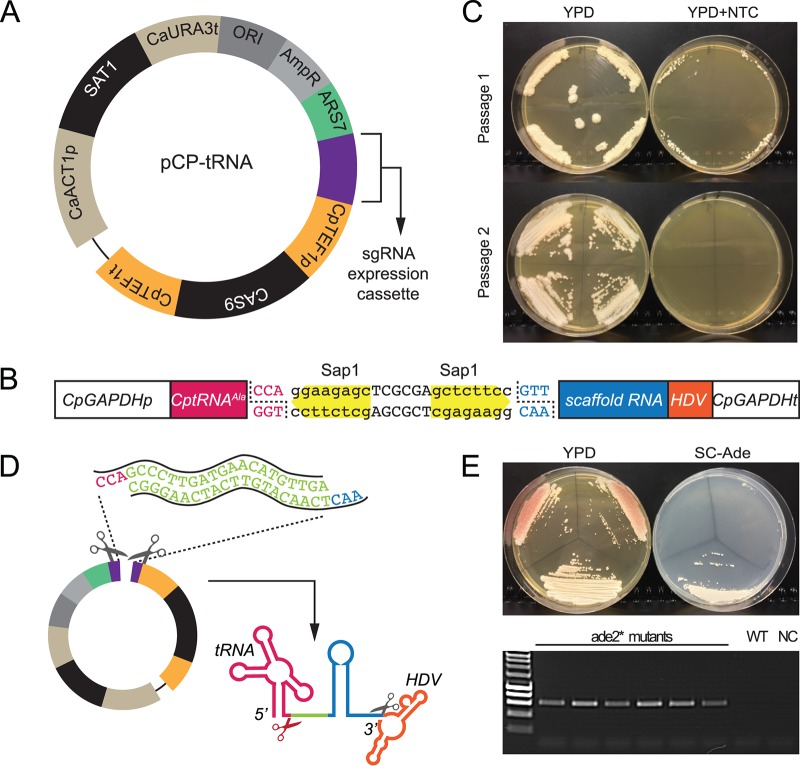
FIG 1.
The pCP-tRNA plasmid system for gene editing in C. parapsilosis. (A) The plasmid shares the main features of the pRIBO system (29), namely, the SAT1 gene (nourseothricin resistance), autonomously replicating sequence 7 (ARS7) from C. parapsilosis, and the CAS9 gene expressed from the C. parapsilosis TEF1 promoter. (B) The pRIBO and pCP-tRNA systems differ in the cassette used to express the sgRNA. In pCP-tRNA, the RNA pol II GAPDH promoter is followed by the tRNAAla sequence (in pink), two SapI restriction sites (in yellow), the scaffold RNA (in blue), and the hepatitis delta virus (HDV) sequence (in orange). (C) Like pRIBO, pCP-tRNA is easily lost. Transformed cells were patched to YPD plates without nourseothricin (NTC) for 48 h and were then streaked on YPD and YPD plus NTC. Colonies from YPD were repatched after 48 h. All transformants lost NTC resistance after just two passages. (D) The target guide (in green, representing ADE2-B in panel E) was generated by annealing two 20-bp oligonucleotides carrying overhang ends (in pink and blue) and cloned into SapI-digested pCP-tRNA. The guide RNA is released by cleavage after the tRNAAla and before the HDV ribozyme. (E) Editing of ADE2 using the pCP-tRNA system. Transformation of C. parapsilosis CLIB214 with pCP–tRNA–ADE2-B and a repair template (RT-B [29]) resulted in the introduction of two stop codons that disrupted the gene function, producing pink colonies that failed to grow in the absence of adenine (SC-ade). A white Ade-positive (Ade+) wild-type colony is shown as a control. The transformants were screened by PCR using the mutADE2B-F primer derived for the edited site and the downstream ADE2_REV primer, which generates a product only when the mutation is present as described in reference 29. WT (wild type), CLIB214 strain; NC, no DNA.