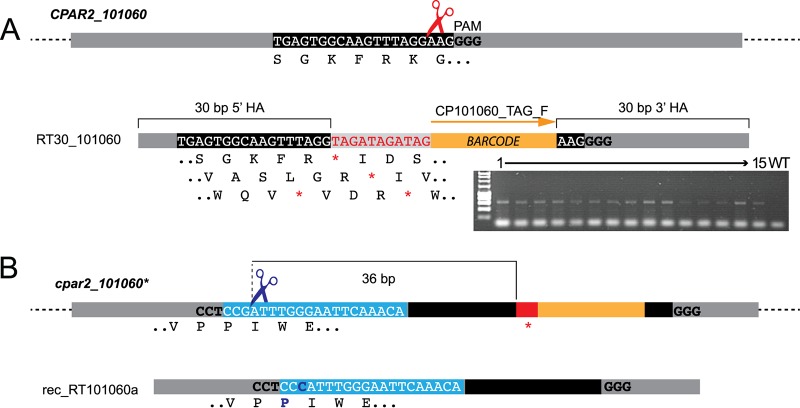
FIG 2.
Editing and reconstitution of CPAR2_101060. (A) The plasmid pCP-tRNA-CP101060 was generated to target CPAR2_101060. The guide sequence recognized by Cas9 is boxed in black, and the PAM is shown in bold. The Cas9 cut site is indicated by red scissors. C. parapsilosis CLIB214 cells were transformed with pCP-tRNA-CP101060 and a repair template (RT30_101060) generated by overlapping PCR using RT30_101060_TOP and RT30_101060_BOT oligonucleotides. The repair template contains two 30-bp homology arms (HA) that flank an 11-bp sequence containing coding stop codons in all three possible reading frames (in red, with all reading frames indicated below the sequence) and a 20-bp unique barcode (in orange). The gel shows results of screening of 15 transformants by PCR using primer CP101060_TAG_F, which anneals to the barcode, together with the CP101060_WT_R downstream primer. Sequencing confirmed that stop codons were introduced into both alleles of CPAR2_101060. (B) To replace the cpar2_101060* edited alleles with wild-type sequences, a PAM site (bold) upstream from the edited site (red) was selected. The guide RNA is boxed in light blue. Transformation with pCP-rec-tRNA-Cp101060a containing this guide resulted in Cas9 cleavage 36 bp upstream from the mutated region (indicated by blue scissors). The repair template (rec-RT101060a) generated by overlapping PCR with primers rec-RT-101060aTOP and rec-RT-101060aBOT was designed to replace the edited site and barcode with wild-type sequences. It also included a single G1420C synonymous SNP so that the reconstituted and wild-type alleles could be distinguished. The wild-type sequence was successfully reintroduced in 2/9 transformants tested. The scheme is not drawn to scale.