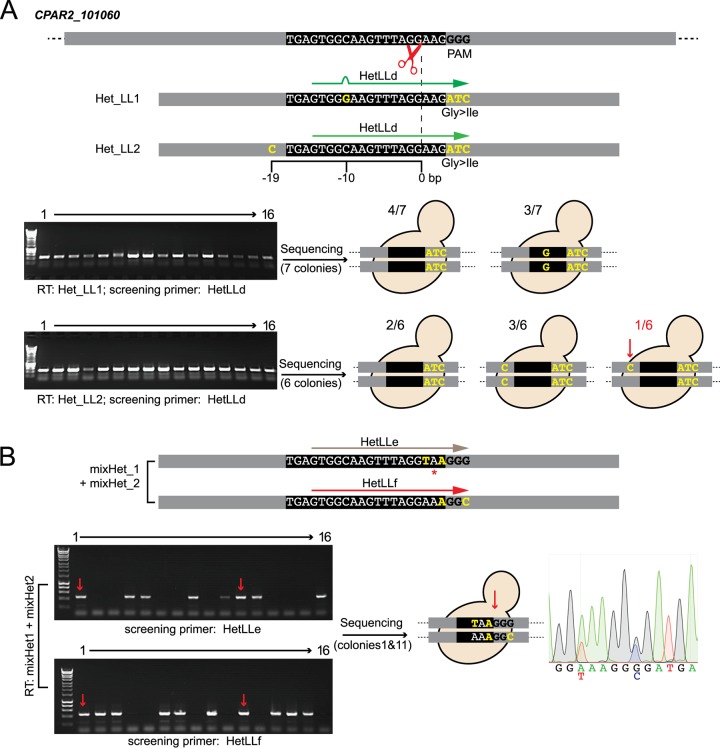
FIG 5.
Generation of heterozygous mutations with the CRISPR-Cas9 system. (A) Varying the distance between the Cas9 cut site and introduced DNA change. (B) Using a mixture of repair templates. (A) C. parapsilosis CLIB214 cells were transformed with pCP-tRNA-CP101060 and either the Het_LL1 repair template or the Het_LL2 repair template. Both Het_LL1 and Het_LL2 carry three nucleotide changes that introduce a codon change and disrupt the PAM (GGG to ATC, resulting in a Gly-to-Ile amino acid change) (yellow). The two templates also carry an additional SNP which introduces a silent mutation at either 10 bp (C>G) or 19 bp (T>C) upstream from the Cas9 cut site. Sixteen transformants obtained with Het_LL1 and Het_LL16 transformed with Het_LL2 yielded a PCR product using primer HetLLd, which anneals at the ATC codon change, and the CP101060_WT_R downstream primer. HetLLd does not anneal perfectly to the sequence introduced by Het_LL1, because of the C>G SNP. The cartoons show the sequencing results. With repair template Het_LL1, all 7 transformants contained the amino acid change at both alleles; in 3 transformants, G was introduced at both alleles 10 bp upstream of the cut site, and four retained the wild-type C base. With repair template Het_LL2, all 6 transformants again contained the amino acid change at both alleles; 5 transformants contained either the wild-type base or the mutated base at the additional site, but one was heterozygous for a T>C SNP 19 bp from the cut site. The combination of alleles in this strain is referred to as CP101060-ATC/CP101060-ATC-SNP. (B) C. parapsilosis CLIB214 was transformed with pCP-tRNA-CP101060 and a mixture of two repair templates, mixHet_1 and mixHet_2. MixHet_2 is designed to introduce two silent mutations that do not change the coding sequence but that do disrupt the PAM site, preventing Cas9 from cutting again at the edited site. mixHet_1 also contains two mutations which change the protospacer preventing the gRNA binding and which introduce a stop codon. Among the 16 transformants, 2 (indicated by the red arrows) generated PCR products when amplified with primers annealing to either of the targeted editing sites (HetLLe or HetLLf) and the CP101060_WT_R downstream primer. Sequencing of these colonies confirmed that different repair templates had been incorporated at each allele. The chromatogram shows one example. Genotype: CP101060-Stop/CP101060-SNP.