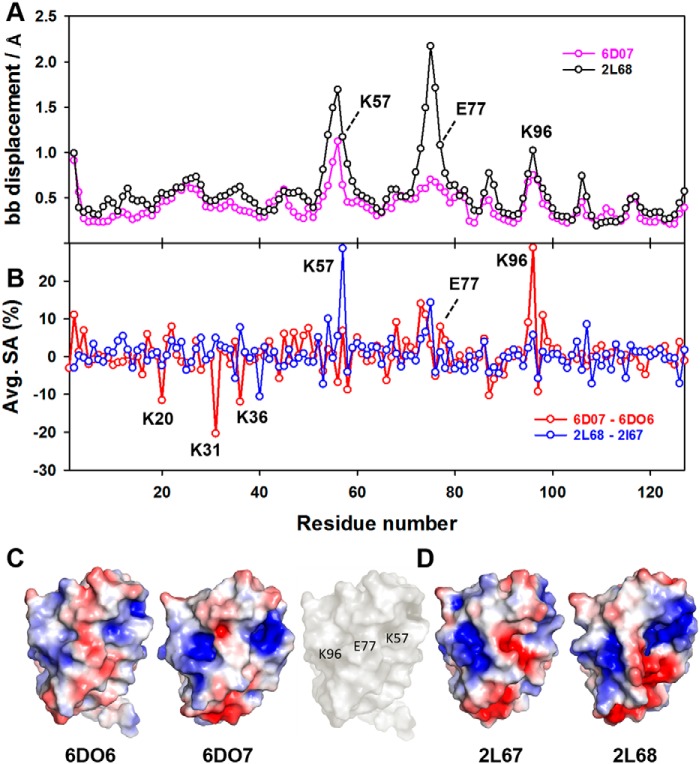
Figure 8.
Solvent accessibility, backbone displacement, and electrostatic surface for FABP1 in the absence and presence of GW7647. A, global backbone displacement between apo and holo NMR structures of FABP1 in the absence and presence of GW7647 (6DO7 versus 6DO6) and oleic acid (2L68 versus 2L67) plotted against residue number. B, change in average solvent accessibility per residue for FABP1 in the absence and presence of GW7647 (6DO7 versus 6DO6) and oleic acid (2L68 versus 2L67) plotted against residue number. Solvent accessibility and backbone displacements were calculated in MOLMOL. C, electrostatic surfaces for apo FABP1 (6DO6) and holo FABP1 bound to GW7647 (6DO7). For 6DO6 and 6DO7, the conformer closest to the mean of the ensemble was used to calculate the electrostatic surface. The surface of apo FABP1 (6DO6) in white showing the position of residues Lys-57, Glu-77, and Lys-96. D, electrostatic surfaces for apo FABP1 (2L67) and holo FABP1 bound to oleic acid (2L68). For 2L67 and 2L68, the first conformer of the ensemble was used to calculate the electrostatic surface. Electrostatic surface potentials were calculated using the APBS plug-in within PyMOL.