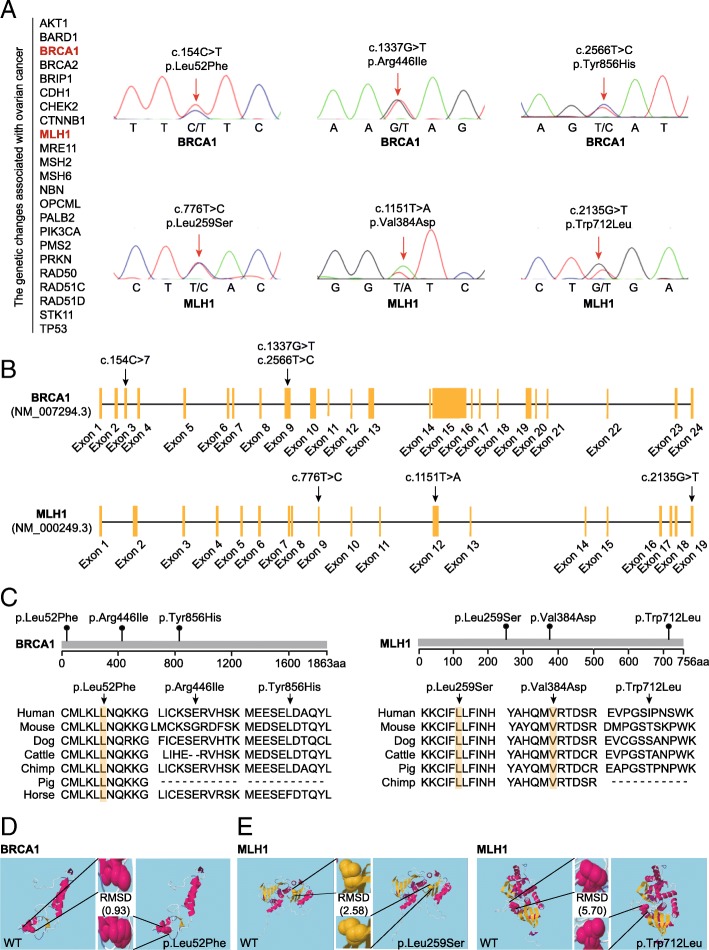
Fig. 4.
Identification of the BRCA1 and MLH1 mutations. a The genetic changes in 23 genes associated with ovarian cancer were analyzed by whole-exome sequencing. Sanger sequencing was performed to confirm the variants detected by whole-exome sequencing. b Locations and conservation of mutations in the BRCA1 and MLH1 genes. The positions of all mutations are indicated in the BRCA1 and MLH1 genomic structures. c Conservation of the mutated amino acids is indicated by the alignment of seven mammalian species. d Secondary structural modeling of the wild-type BRCA1 and the p.Leu52Phe variant. e Secondary structural modeling of the wild-type MLH1, and the p.Leu259Ser and p.Trp712Leu variants. Modeling was performed using RaptorX (http://raptorx.uchicago.edu/). RMSD, root mean square deviation, which represents the average distance between atoms of the wild-type and mutant proteins