Figure 4.
Loss of Sfpq Downregulates Energy Metabolism Pathway Gene Expression in Muscle Cells
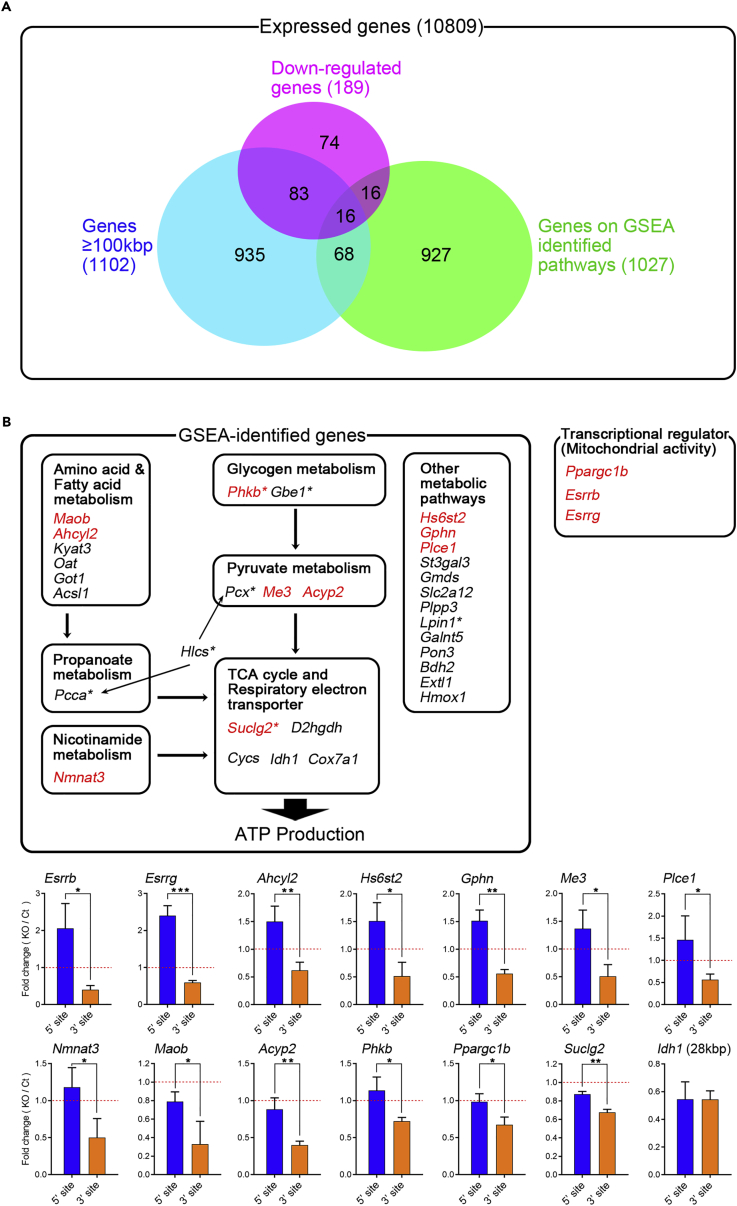
(A) Venn diagram of downregulated genes, genes≥100 kbp, and genes on GSEA identified pathways.
(B) Upper panel: Schematic diagram of downregulated genes in KO myotubes, which are GSEA-identified metabolic-pathway-related genes and also transcriptional regulators for mitochondrial activity. Asterisk indicates HGMD-identified inherited metabolic-diseases-related genes in Figure S2E. Lower panel: RT-qPCR for 5′ and 3′ sites of pre-mRNAs (n = 3). Pre-mRNA ratio (fold change) was calculated between KO and control myotubes for each position (Control = 1). Idh1 gene was used as negative control (the gene length <100 kbp). Genes showing 3′-site downregulation are highlighted with red characters in upper panel. Data are presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 (Student's or Welch's t test).
See also Figures S2–S4.