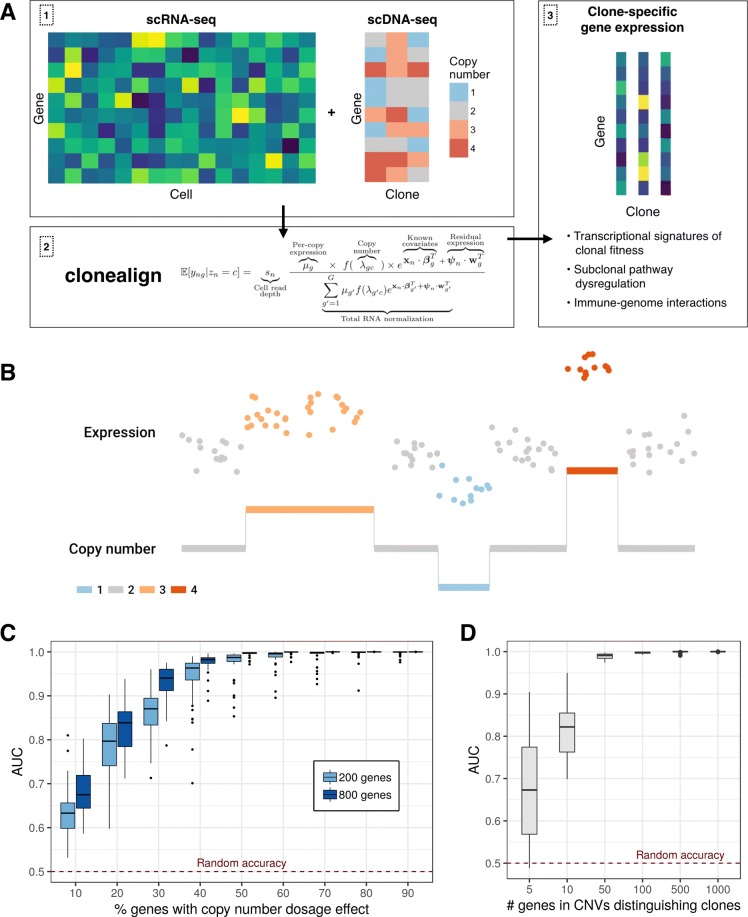
Fig. 1.
Assigning single-cell RNA-seq to clone-of-origin using clonealign. a Given independently sampled single-cell DNA- and RNA-seq from the same tumor, the clonealign statistical framework assigns each cell’s gene expression profile to its clone-of-origin, uncovering transcriptional signatures of clonal fitness. b To relate cells as measured in RNA-space to their clones measured in DNA-space, we assume a relationship between gene copy number and gene expression (simulated data). c Simulations demonstrate the robustness of clonealign to the underlying proportion of genes exhibiting a copy number dosage effect. Even if only 30% of genes have a clone-specific copy number effect on expression, clones can still be accurately assigned with an average AUC >0.8. d Simulations demonstrate clonal assignment is accurate even when as few as 10–50 genes lie in regions of differing copy number between clones, allowing clonal assignment from only small-scale genomic rearrangements