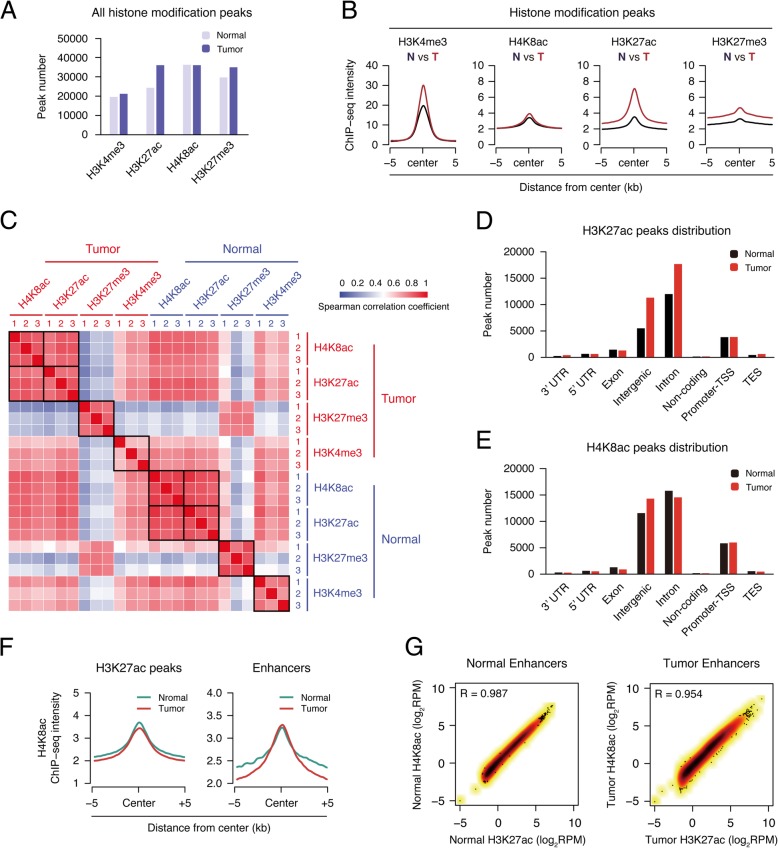
Fig. 2.
H4K8ac has a high correlation with H3K27ac in the position of the enhancer. a Bar plot showing the number of significant peaks from four histone modifications (H3K4me3, H3K27ac, H4K8ac, and H3K27me3) in both normal tissue and tumor. b Metagene plot representation of the mean ChIP-seq signal for four indicated histone marks across the individual peaks of each mark. Metagene analysis is centered on the middle of peaks and 10 kb around the center of peaks are displayed (5 kb upstream and 5 kb downstream). Each ChIP-seq data of histone marks were merged by their three individual replicates. c Heat map representation of correlation based on four histone modifications (H3K4me3, H3K27ac, H4K8ac, and H3K27me3) occupancy at mouse genome wide. Sample correlations were calculated by Spearman correlation coefficient. d, e Distribution of histone mark H4K8ac and H3K27ac in different genome elements according to the location of their peaks. Promoter-TSS, “− 1kb to + 100bp” of the transcription start sites. TES, “− 100bp to + 1Kb” of transcription termination sites. f Metagene plot representation of the mean H4K8ac signal across H3K27ac peaks and enhancer in both normal tissue and tumor. g Scatter plots showing the correlation of H3K27ac and H4K8ac levels in both normal tissue and tumor enhancer. Correlation was calculated by the Pearson correlation coefficient