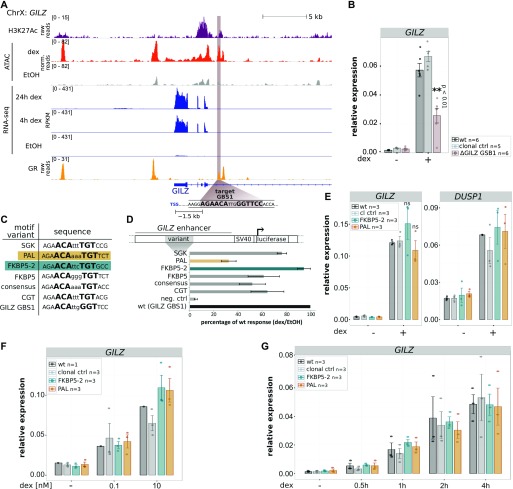
Figure 5. Effect of GBS1 sequence identity on GR-dependent GILZ activation.
(A) Tracks showing H3K27ac and GR ChIP-seq tag density, ATAC-seq, and RNA-seq reads at the GILZ locus for U2OS-GR cells treated as indicated. The GILZ GBS1 targeted for editing is highlighted in brown and the distance in kilo base pairs (kb) to the next TSS is indicated. (B) Relative GILZ mRNA expression is shown for parental U2OS-GR cells, for unedited clonal controls, and for clonal lines with a deleted GBS1. The average ± SEM of at least five clonal cell lines treated overnight with 1 μM dex or vehicle control (−) is shown. Dots show the values for each individual clonal line. Statistical tests were performed using an unpaired two-sided Mann–Whitney U test comparing dex-treated GILZ levels between clonal lines with a deleted GBS1 and their unedited clonal control counterpart. (C) DNA sequence of GBS variants analyzed. (D) Relative fold activation of luciferase reporters with GBS variants as indicated comparing cells treated with vehicle control (etoh) and cells treated overnight with 1 μM dex. Averages ± SEM from three independent experiments are shown. (E) Relative mRNA expression levels as determined by qPCR for GILZ and for the unedited control GR target gene DUSP1 are shown for unedited parental U2OS-GR cells (wt), for unedited clonal control cell lines, and for clonal cell lines with GBS variant as indicated at the GILZ GBS1 locus. Averages ± SEM for cell lines treated overnight with 1 μM dexamethasone (dex) or with ethanol (−) as vehicle control are shown. Dots show the values for each individual clonal line. Statistical tests were performed using an unpaired two-sided Mann–Whitney U test comparing dex-treated GILZ levels between clonal lines for each introduced GBS variant and their unedited clonal control counterpart. (E, F) Same as for (E) except that GILZ mRNA levels are shown for cells treated overnight with 0.1 nM dex, 10 nM dex, or vehicle control (−). (E, G) Same as for (E) except that cells were treated for 0.5, 1, 2, or 4 h with 1 μM dex.