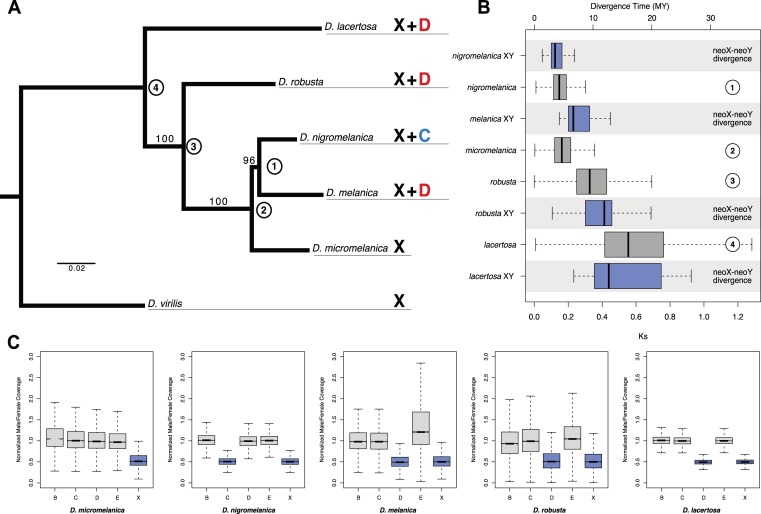
Fig 2. Phylogenetic relationships and karyotype evolution in the D. melanica and D. robusta species groups.
(A) Whole-genome maximum likelihood phylogeny inferred using the RAxML rapid bootstrapping algorithm. Bootstrap support values are shown on branches, and nodes are numbered for reference to Panel B. Each species is labeled according to which chromosomes are X linked. X refers to the ancestral X chromosome, while the other chromosomes are referred to as Muller elements B–E (the small dot chromosome, Muller F, is not shown). (B) Divergence (calculated as Ks and converted to MY ago) between neo-X/Y chromosomes as well as the nodes labeled in Panel A. Boxes reflect the distribution of Ks values for all neo-X/Y gene pairs or between-species orthologs. Underlying data can be found in S1 Data. (C) X-linkage was determined based on the ratio of male/female Illumina sequencing coverage for genomic libraries aligned to the female assemblies (autosomes = gray, X chromosomes = blue). Underlying data can be found in S2 Data. MY, million years.