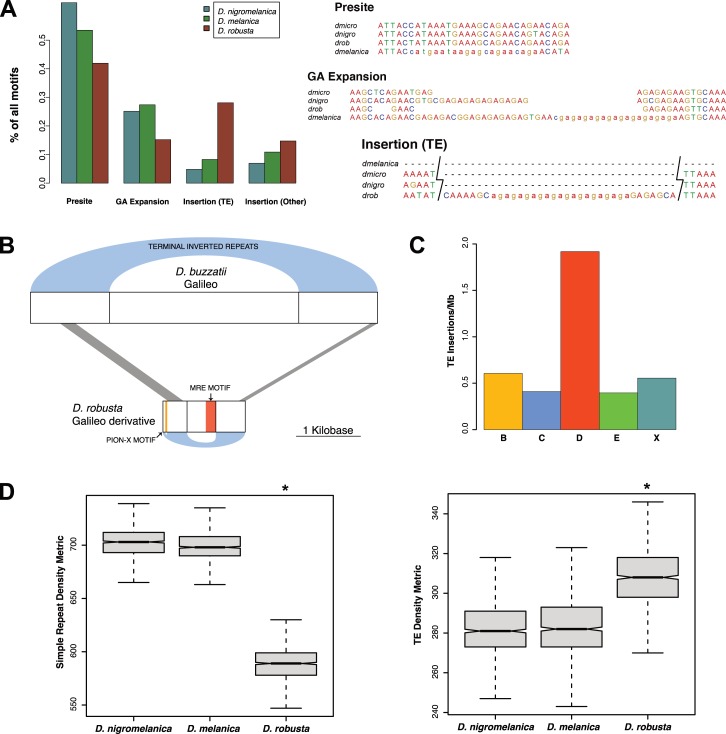
Fig 5. Evolution of CES motifs.
(A) We used a comparative genomics approach to determine the evolutionary mechanism that gave rise to the MSL-binding motifs found within each CES on the neo-X chromosome of each species; shown is one example of each major type. D. robusta has a larger number of CES motifs derived from TE insertions compared to the other two species, whereas D. nigromelanica and D. melanica have a larger number of motifs that arose from GA expansion. Underlying data can be found in S5 Data. (B) We identified a Galileo-like TE that gave rise to at least 24 CES motifs on the D. robusta neo-X chromosome. (C) Insertions of this TE are significantly enriched on the D. robusta neo-X, compared to both autosomes and the ancestral X chromosome (binomial test P = 3.2 × 10‐14). Underlying data can be found in S5 Data. (D) Both D. nigromelanica and D. melanica genome assemblies have a higher density of simple repeats, whereas the D. robusta genome assembly has a higher density of TEs (Wilcoxon test P < 2.2 × 10‐16 for both comparisons). The repeat density metric refers to the number of 1-kb windows that overlap a repeat in the genome assembly out of 1,000 randomly placed windows. Underlying data can be found in S5 Data. CES, chromatin entry site; MRE, MSL recognition element; MSL, male-specific lethal; pion-X site, pioneering site on the X; TE, transposable element.