Extended Data Fig. 8 |. The zinc-finger domains of NEIL3 contribute to its recruitment to the replication fork.
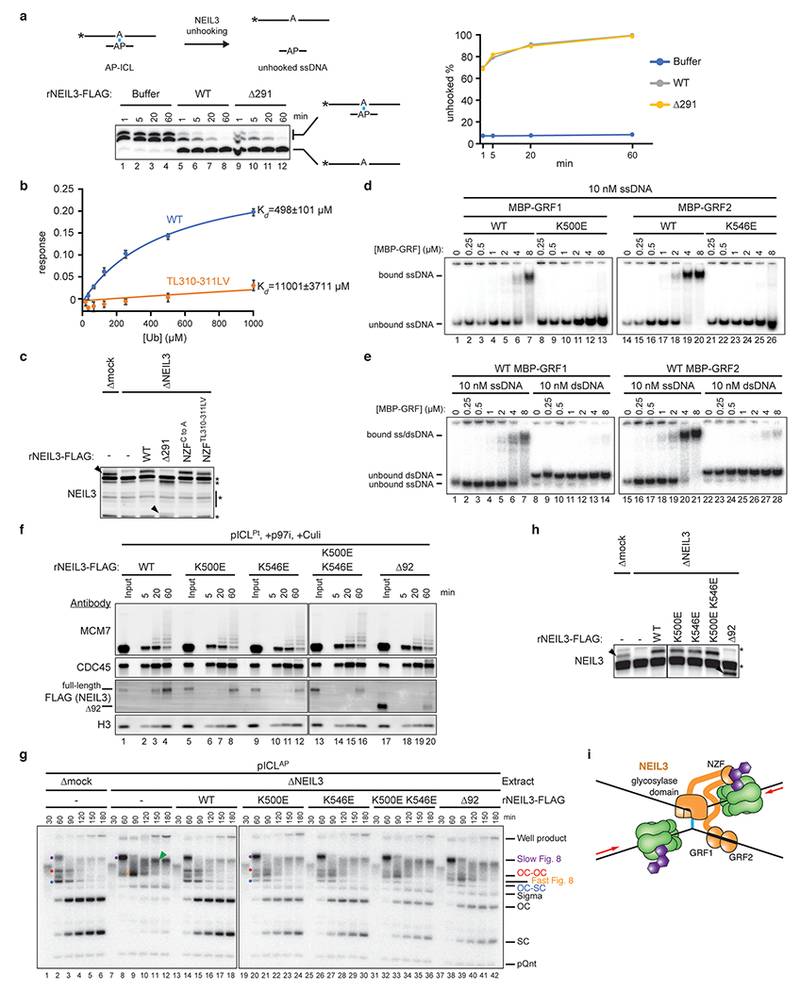
a, Left, to determine whether rNEIL3Δ291 is catalytically active, a model AP-ICL substrate comprising a synthetic 5′-radiolabeled 24mer oligonucleotide cross-linked to a ~3mer was mixed with rNEIL3Δ291 or rNEIL3WT. Cross-linked and unhooked species were resolved by denaturing polyacrylamide gel electrophoresis and visualized by autoradiography. Asterisks indicate the 32P radiolabel. Note that the cross-linked species migrates as a doublet due to heterogeneity in the bottom strand following RNase digestion (see Methods for details). Right, quantification of unhooking. Equivalent results were obtained in three independent experiments, which show that rNEIL3Δ291 retains full glycosylase activity.
b, Interaction of the NEIL3 NPL4-type zinc finger (NZF; residues 300 to 328) with ubiquitin. GST-NEIL3 NZF fusion protein (WT or TL,LV substituted) was immobilized on a biosensor tip and monoubiquitin binding was measured by biolayer interferometry (BLI). The ubiquitin binding response was corrected for non-specific binding to GST and plotted as a function of ubiquitin concentration. Error bars represent standard error of the mean from three independent experiments.
c, Extracts used in Fig. 3e were blotted for NEIL3. Black arrowheads, NEIL3-specific bands. rNEIL3Δ291 is not efficiently detected by the NEIL3-specific primary antibody. Non-specifically detected proteins are marked with asterisks.
d and e, To test whether the two GRF zinc fingers in NEIL3 interact with ssDNA, we expressed each individually and performed electrophoretic mobility shift assays. rMBP-NEIL3 GRF zinc finger fusion proteins (wild-type or substituted) were incubated with 5′-radiolabeled 25-mer ssDNA or dsDNA. Bound and unbound DNAs were resolved by native polyacrylamide gel electrophoresis and visualized by autoradiography. This analysis reveals that both GRF domains bind specifically to ssDNA.
f, Analysis of proteins associated with pICLPt during replication in undepleted extract in the presence of p97i and Culi. Extracts were supplemented with wild-type or mutated rNEIL3. At different times, chromatin was recovered and blotted for the indicated proteins. The individual GRF substitutions modestly affected recovery of rNEIL3 upon pICL pull-down while combination of the substitutions or deletion of both GRF zinc fingers strongly reduced rNEIL3 recovery, indicating that interactions mediated by the GRF zinc fingers promote recruitment of NEIL3 to an ICL.
g, pICLAP was replicated in mock- or NEIL3-depleted extracts supplemented with wild-type or mutated NEIL3 as indicated and analyzed as in Fig. 1b. Relative to rNEIL3WT, rNEIL3 with substitutions in either GRF zinc finger that abolish ssDNA binding (K500E and K546E) exhibited modest defects in pICLAP unhooking that were exacerbated when the substitutions were combined, indicating that interactions between the GRF zinc fingers and ssDNA contribute to ICL repair.
h, Extracts used in the replication reactions shown in g, were blotted for NEIL3. Non-specifically detected proteins are marked with asterisks.
i, Model for recruitment of NEIL3 to chromatin by zinc finger-mediated interactions. Upon replication fork convergence at an ICL, TRAIP-dependent CMG ubiquitylation recruits NEIL3 through direct interactions between NEIL3’s NZF domain and ubiquitin. Association of NEIL3 with chromatin is further enhanced by interactions between the tandem GRF zinc fingers and single stranded DNA, possibly on the lagging strand template.
