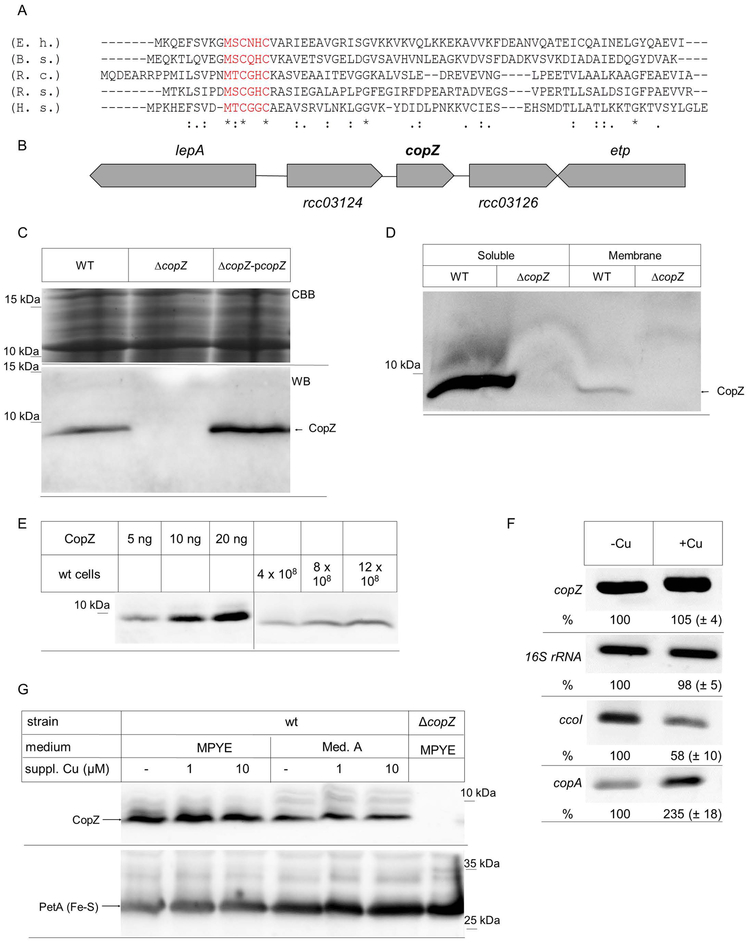
Figure 1: Identification of a CopZ homologue in R. capsulatus.
(A) Amino acid alignment of the R. capsulatus CopZ (R.c.) with the respective CopZ-homologues of Enterococcus hirae (E.h.) Bacillus subtilis (B.s.), Rhodobacter sphaeroides (R.s.), and Homo sapiens (H.s.). The conserved Cu-binding motif is shown in red. (B) Genetic organization of copZ in R. capsulatus. lepA and etp presumably encode for a translation factor and a phosphotyrosine protein phosphatase, respectively. The open reading frames rcc03124 and rcc03126 encode for hypothetical proteins. (C) R. capsulatus cells were grown in MPYE medium, precipitated with trichloroacetic acid and the pellet was dissolved loading buffer. After SDS-PAGE, the gel was either stained directly with coomassie brilliant blue (CBB) as loading control, or was blotted and decorated with α-CopZ antibodies (WB). WT corresponds to MT1131, ΔcopZ to a MT1131 derivative carrying an insertion-deletion mutation within copZ and ΔcopZ-pcopZ to the ΔcopZ strain with a plasmid-encoded copZ. (D) MT1131 (WT) and ΔcopZ cell extracts were separated into a soluble fraction and a membrane fraction by ultracentrifugation. Subsequently, the material was separated by SDS-PAGE and decorated with α-CopZ antibodies. (E) The cellular concentration of CopZ in MT1131 grown on MPYE medium without further Cu supplementation was determined by quantitative western blotting, using defined amounts of purified CopZ as reference. Signal intensity was quantified by ImageJ and several independent experiments were performed and a representative western blot is shown. Note that the purified CopZ contained a His-tag and it therefore migrates slower on SDS-PAGE than the native CopZ. (F) RT-PCR analyses of mRNA levels in wild type cells grown on MPYE without and with Cu supplementation (10 μM Cu(II)). A representative gel of three independent experiments is shown. The 16S ribosomal RNA served as control and the ccoI and copA mRNA as reference. Quantification was performed with ImageJ and signal intensity of the mRNA level in cells without Cu supplementation was set to 100%. (G) The CopZ levels in whole cells grown either on enriched medium (MPYE) or minimal medium (MedA) were analysed by immunoblotting as described above. When indicated, CuSO4 was added to the growth medium. The levels of the Rieske Fe-S protein PetA served as loading control.