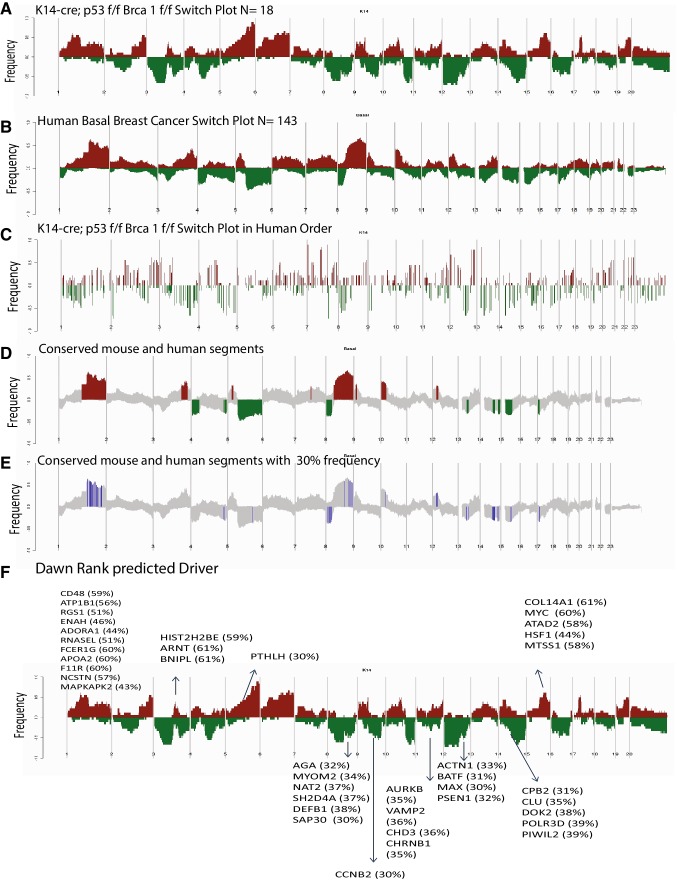
Fig. 3.
Analysis of K14-Cre; p53f/f Brca1f/f tumors reveals conserved copy number alterations with basal-like human breast cancer. a Gains (red) and losses (green) are shown for the KPB1 murine tumors, b human basal breast tumors, c the KPB1 murine tumors with gains/losses shown in human chromosome order. d All conserved gains/losses between murine KPB1 tumors and human basal-like tumors are shown. e Conserved gains and losses with a 30% or greater frequency in both KPB1 and human basal-like tumors are shown. f The mapping of driver mutations predicted by the DawnRank algorithm is shown on the KPB1 murine switch plot from a. For all panels, the frequency of alterations in each group is indicated on the y-axis, where the frequency of losses is shown from 0.0 to − 1.0 (for example, a value of − 0.5 indicates loss in 50% of samples) and frequency of gains is shown from 0.0 to 1.0