Figure 6.
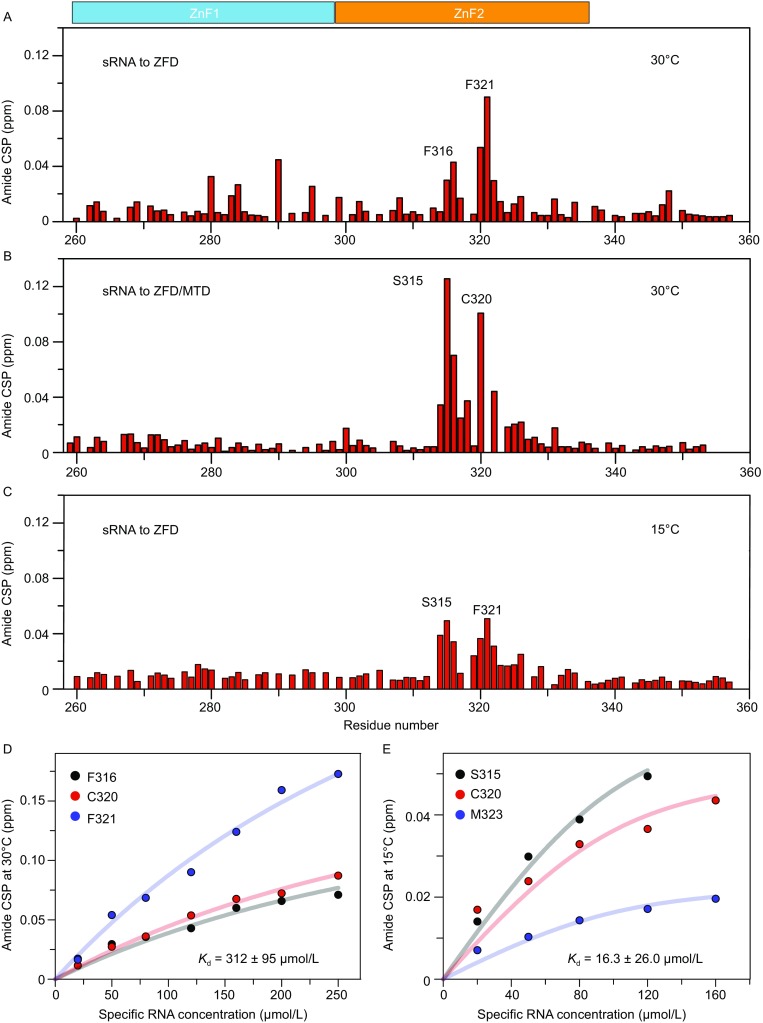
NMR titrations for METTL ZFD with the specific RNA substrate. (A and C) Chemical shift perturbations (CSPs) for the backbone amide of 100 µmol/L 15N-labeled ZFD mixed with 120 µmol/L 20-nt RNA substrate containing the m6A consensus sequence at two different temperatures. Note the NMR experiment could not be performed at 10°C due to severe line broadening of the free protein. No residues disappear upon RNA titration when RNA concetration is lower than 120 μM. Proline, residues with severe signal overlapping or residues without assignment is indicated with a gap in the histogram. (B) The CSPs for 15N-labeled ZFD natively ligated to the MTD heterodimer of METTL3-METTL14, upon the titration of specific RNA. The experimental conditions are the same as in panel A except for the protein used. Residues F321 and M323 disappeared in the complex, precluding accurate assessment of their CSPs. (D and E) The Kd values for the interactions between ZFD and RNA obtained by fitting the CSPs at the two temperatures. Note that molar ratio of RNA relative to ZFD could only reach ~1.6 at the lower temperature
